
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 1,277,773 – 1,277,870 |
| Length | 97 |
| Max. P | 0.769906 |

| Location | 1,277,773 – 1,277,866 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 63.06 |
| Shannon entropy | 0.62333 |
| G+C content | 0.38307 |
| Mean single sequence MFE | -20.93 |
| Consensus MFE | -7.21 |
| Energy contribution | -6.58 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.55 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.519543 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
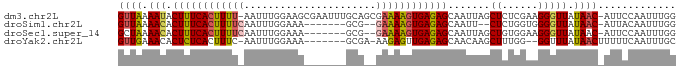
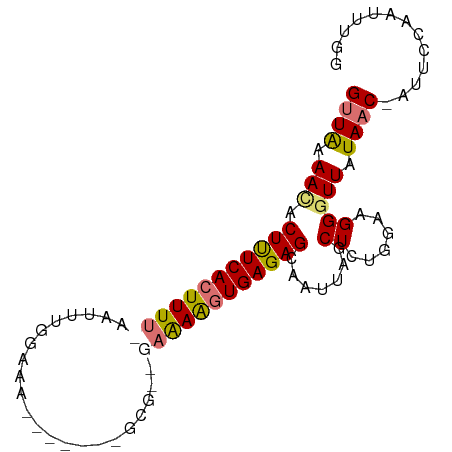
>dm3.chr2L 1277773 93 - 23011544 -------AAAUACUUUCACUUUU-AAUUUGGAAGCGAAUUUGCAGCGAAAAGUGAGAGCAAUUAGCUCUCGAAGGGUUAUAAC-AUUCCAAUUUGGAGCAUU- -------.....(((((((((((-((((((....)))))).......))))))((((((.....)))))))))))........-.((((.....))))....- ( -22.81, z-score = -2.05, R) >droSim1.chr2L 1250403 83 - 22036055 -------AAACACUUUCACUUUUCAAUUUGGAAA-------GCG--GAAAAGUGAGAGCAAUU--CUCUGGUGGGGUUAUAAC-AUUACAAUUUGGAGCAUU- -------.....((((((((((((...((....)-------)..--)))))))))))).(((.--(((..(((..((......-))..))..)..))).)))- ( -18.80, z-score = -1.88, R) >droSec1.super_14 1228260 85 - 2068291 -------AAACACUUUCACUUUUCAAUUUGGAAA-------GCG--GAAAAGUGAGAGCAAUUAGCUGUGGAAGGGUUAUAAC-AUUCCAAUUUGGAGCAUU- -------.....((((((((((((...((....)-------)..--))))))))))))......(((.(((((..((....))-.)))))......)))...- ( -21.60, z-score = -2.28, R) >droYak2.chr2L 1253674 84 - 22324452 -------AAACACUCUCACUUUC-AAUUUGGAAA-------GCGA-AAGAGUUGAGAGCAACAAGCUUUGG--GGUUUAUAACUUUUUCAAUUUGCAACAUU- -------...........(((((-......))))-------).((-((((((((.((((..((.....)).--.)))).)))))))))).............- ( -18.60, z-score = -1.91, R) >dp4.chr4_group3 9752244 93 - 11692001 -GGAGGGAAACACUUUUCCUUUC--GUUUGGAAAAG----UGAAACAGGUGGCAAAAGCAACAGG--AUGGGCACAUUUUACUCUUUGGAUUUGCGGAAAGU- -((((((...(((((((((....--....)))))))----))......((.((....)).))(((--(((....)))))).))))))...............- ( -22.10, z-score = -0.63, R) >droPer1.super_8 952356 95 - 3966273 GGGGGGGAAACACUUUUCCUUUC--GUUUGGAAAAG----UGAAACAGGUGGCAAAAGCAACAGG--AUGGGCACAUUUUACUCUUUGGAUUUGGGGGAAAUU .((((((...(((((((((....--....)))))))----))......((.((....)).))(((--(((....)))))).))))))................ ( -21.70, z-score = -0.48, R) >consensus _______AAACACUUUCACUUUC_AAUUUGGAAA_______GCAA_AAAAAGUGAGAGCAACAAGCUAUGGAAGGGUUAUAAC_AUUCCAAUUUGGAGCAUU_ ............(((((((((((........................)))))))))))............................................. ( -7.21 = -6.58 + -0.64)



| Location | 1,277,779 – 1,277,870 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 78.64 |
| Shannon entropy | 0.33565 |
| G+C content | 0.36226 |
| Mean single sequence MFE | -21.58 |
| Consensus MFE | -9.86 |
| Energy contribution | -11.18 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.769906 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 1277779 91 - 23011544 GUUAAAAUACUUUCACUUUU-AAUUUGGAAGCGAAUUUGCAGCGAAAAGUGAGAGCAAUUAGCUCUCGAAGGGUUAUAAC-AUUCCAAUUUGG ((((.(((.(((((((((((-((((((....)))))).......))))))((((((.....)))))))))))))).))))-............ ( -22.61, z-score = -2.45, R) >droSim1.chr2L 1250409 81 - 22036055 GUUAAAACACUUUCACUUUUCAAUUUGGAAA-------GCG--GAAAAGUGAGAGCAAUU--CUCUGGUGGGGUUAUAAC-AUUACAAUUUGG ((((.(((.((((((((((((...((....)-------)..--)))))))))))).....--(((....)))))).))))-............ ( -20.80, z-score = -3.21, R) >droSec1.super_14 1228266 83 - 2068291 GCUAAAACACUUUCACUUUUCAAUUUGGAAA-------GCG--GAAAAGUGAGAGCAAUUAGCUGUGGAAGGGUUAUAAC-AUUCCAAUUUGG (((((....((((((((((((...((....)-------)..--))))))))))))...)))))..(((((..((....))-.)))))...... ( -24.30, z-score = -3.50, R) >droYak2.chr2L 1253680 82 - 22324452 GUUGAAACACUCUCACUUUC-AAUUUGGAAA-------GCGA-AAGAGUUGAGAGCAACAAGCUUUGG--GGUUUAUAACUUUUUCAAUUUGC ...............(((((-......))))-------).((-((((((((.((((..((.....)).--.)))).))))))))))....... ( -18.60, z-score = -1.10, R) >consensus GUUAAAACACUUUCACUUUU_AAUUUGGAAA_______GCG__GAAAAGUGAGAGCAAUUAGCUCUGGAAGGGUUAUAAC_AUUCCAAUUUGG (((((....((((((((((((......................))))))))))))...))))).............................. ( -9.86 = -11.18 + 1.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:08:11 2011