| Sequence ID | dm3.chr2L |
|---|---|
| Location | 14,066,744 – 14,066,841 |
| Length | 97 |
| Max. P | 0.999636 |

| Location | 14,066,744 – 14,066,841 |
|---|---|
| Length | 97 |
| Sequences | 8 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 62.04 |
| Shannon entropy | 0.73835 |
| G+C content | 0.46898 |
| Mean single sequence MFE | -21.86 |
| Consensus MFE | -10.10 |
| Energy contribution | -9.91 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.31 |
| Mean z-score | -0.91 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.879112 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
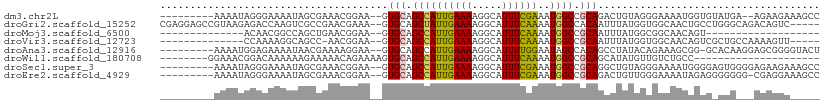
>dm3.chr2L 14066744 97 + 23011544 ---------AAAAUAGGGAAAAUAGCGAAACGGAA--GUGCAGCCAUUGAAAAGGCAUUUCGAAAUGGCCGCAGACUGUAGGGAAAAUGGUGUAUGA--AGAAGAAAGCC ---------...((((.............((....--))((.((((((((((.....))))))..)))).))...))))..................--........... ( -15.80, z-score = 0.09, R) >droGri2.scaffold_15252 4582317 103 + 17193109 CGAGGAGCCGUAAGAGACCAAGUCGCCGAACGAAA--GUGCAGCUAUUGAAAAGGCAUUUCAAAAUGGCCACAAUUUAUGGUGGCAACUGCCUGGGCAGACAGUC----- .((((.(((......(((...)))((.(.((....--)).).)).........))).))))......(((((........)))))..((((....))))......----- ( -29.40, z-score = -1.03, R) >droMoj3.scaffold_6500 22140945 75 - 32352404 --------------ACAACGGCCAGCUGAACGGAA--GUGCAGCCAUUGAAAAGGCAUUUCAAAAUGGCCGCAAUUUAUGGCGGCAACAGU------------------- --------------....((((((((((.((....--)).))))..((((((.....))))))..))))))...........(....)...------------------- ( -24.50, z-score = -1.89, R) >droVir3.scaffold_12723 1163987 88 + 5802038 --------------CCAAAAGGCAGCC-AACGGAA--GUGCAGCCAUUGAAAAGGCAUUUCAAAAUGGCCGCAAUUUAUGGUGGCAACAGUCGCUGCCAAAAGUU----- --------------......((((((.-.((....--))((.((((((((((.....)))))..))))).)).......((((....)).)))))))).......----- ( -28.60, z-score = -1.81, R) >droAna3.scaffold_12916 4466785 98 - 16180835 ---------AAAAUGGAGAAAAUAACGAAAAGGAA--GUGCAGCCAUUGAAAAGGCAUUUUGGAAUAGCCACAGCCUAUACAGAAAGCGG-GCACAAGGAGCGGGGUACU ---------.........................(--((((.(((.(((....((((((....))).)))...((((...........))-)).))).).))...))))) ( -17.10, z-score = 0.25, R) >droWil1.scaffold_180708 11111830 81 - 12563649 --------GGAAACGGACAAAAAAGAAAAACAGAAAAGUGCAGCCAUUGAAAAGGCAUUUCAAAAUGGCCGCAGCAUAUGUUGUCUGCC--------------------- --------.....(((((((.........((......))((.((((((((((.....)))))..))))).))........)))))))..--------------------- ( -18.50, z-score = -1.35, R) >droSec1.super_3 324863 99 + 7220098 ---------AAAAUAGGGAAAAUAGCGAAACGGAA--GUGCAGCCAUUGAAAAGGCAUUUCGAAAUGGCCGCAGGCUGUAGGGAAAAUGGGGAGUGGGGAGAAGAAAGCC ---------...........(((.((...((....--))...)).))).....(((.((((.......((.((..((.((.......)).))..)).))....))))))) ( -19.40, z-score = -0.45, R) >droEre2.scaffold_4929 15238744 98 - 26641161 ---------AAAAUAGGGAAAAUAGCGAAACGGAA--GUGCAGCCAUUGAAAAGGCAUUUCGAAAUGGCCGCAGACUGUUGGGAAAAUAGAGGGGGGG-CGAGGAAAGCC ---------...........(((.((...((....--))...)).))).....(((.((((....(.(((.(...(((((.....)))))....).))-).).))))))) ( -21.60, z-score = -1.12, R) >consensus _________AAAAUAGAGAAAAUAGCGAAACGGAA__GUGCAGCCAUUGAAAAGGCAUUUCAAAAUGGCCGCAGUCUAUGGGGAAAACAGUG_GUG___AGAGGA_____ ......................................(((.((((((((((.....))))))..)))).)))..................................... (-10.10 = -9.91 + -0.19)

| Location | 14,066,744 – 14,066,841 |
|---|---|
| Length | 97 |
| Sequences | 8 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 62.04 |
| Shannon entropy | 0.73835 |
| G+C content | 0.46898 |
| Mean single sequence MFE | -21.38 |
| Consensus MFE | -14.39 |
| Energy contribution | -14.36 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.11 |
| SVM RNA-class probability | 0.999636 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
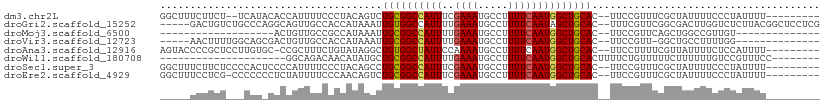
>dm3.chr2L 14066744 97 - 23011544 GGCUUUCUUCU--UCAUACACCAUUUUCCCUACAGUCUGCGGCCAUUUCGAAAUGCCUUUUCAAUGGCUGCAC--UUCCGUUUCGCUAUUUUCCCUAUUUU--------- ...........--........................((((((((((..((((.....)))))))))))))).--..........................--------- ( -16.30, z-score = -2.16, R) >droGri2.scaffold_15252 4582317 103 - 17193109 -----GACUGUCUGCCCAGGCAGUUGCCACCAUAAAUUGUGGCCAUUUUGAAAUGCCUUUUCAAUAGCUGCAC--UUUCGUUCGGCGACUUGGUCUCUUACGGCUCCUCG -----((((((((....))))))))(((..((.....)).(((((..((((((.....))))))..((((.((--....)).))))....)))))......)))...... ( -28.40, z-score = -1.50, R) >droMoj3.scaffold_6500 22140945 75 + 32352404 -------------------ACUGUUGCCGCCAUAAAUUGCGGCCAUUUUGAAAUGCCUUUUCAAUGGCUGCAC--UUCCGUUCAGCUGGCCGUUGU-------------- -------------------...................((((((...((((((.....)))))).(((((.((--....)).)))))))))))...-------------- ( -21.80, z-score = -1.22, R) >droVir3.scaffold_12723 1163987 88 - 5802038 -----AACUUUUGGCAGCGACUGUUGCCACCAUAAAUUGCGGCCAUUUUGAAAUGCCUUUUCAAUGGCUGCAC--UUCCGUU-GGCUGCCUUUUGG-------------- -----......(((((((....)))))))(((.....(((((((((..(((((.....)))))))))))))).--......)-))...........-------------- ( -28.50, z-score = -2.34, R) >droAna3.scaffold_12916 4466785 98 + 16180835 AGUACCCCGCUCCUUGUGC-CCGCUUUCUGUAUAGGCUGUGGCUAUUCCAAAAUGCCUUUUCAAUGGCUGCAC--UUCCUUUUCGUUAUUUUCUCCAUUUU--------- ...............((((-..(((...((...((((..(((.....)))....))))...))..))).))))--..........................--------- ( -17.20, z-score = -1.07, R) >droWil1.scaffold_180708 11111830 81 + 12563649 ---------------------GGCAGACAACAUAUGCUGCGGCCAUUUUGAAAUGCCUUUUCAAUGGCUGCACUUUUCUGUUUUUCUUUUUUGUCCGUUUCC-------- ---------------------(((.(((((........((((((((..(((((.....)))))))))))))((......)).........))))).)))...-------- ( -20.60, z-score = -2.43, R) >droSec1.super_3 324863 99 - 7220098 GGCUUUCUUCUCCCCACUCCCCAUUUUCCCUACAGCCUGCGGCCAUUUCGAAAUGCCUUUUCAAUGGCUGCAC--UUCCGUUUCGCUAUUUUCCCUAUUUU--------- ((((.............................))))((((((((((..((((.....)))))))))))))).--..........................--------- ( -18.75, z-score = -2.83, R) >droEre2.scaffold_4929 15238744 98 + 26641161 GGCUUUCCUCG-CCCCCCCUCUAUUUUCCCAACAGUCUGCGGCCAUUUCGAAAUGCCUUUUCAAUGGCUGCAC--UUCCGUUUCGCUAUUUUCCCUAUUUU--------- (((.......)-))................(((.(..((((((((((..((((.....)))))))))))))).--..).)))...................--------- ( -19.50, z-score = -2.80, R) >consensus _____UCCUCU___CAC_CACCGUUGUCACCAUAGACUGCGGCCAUUUCGAAAUGCCUUUUCAAUGGCUGCAC__UUCCGUUUCGCUAUUUUCUCUAUUUU_________ .....................................((((((((((..((((.....))))))))))))))...................................... (-14.39 = -14.36 + -0.03)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:39:15 2011