
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 14,045,808 – 14,045,905 |
| Length | 97 |
| Max. P | 0.984633 |

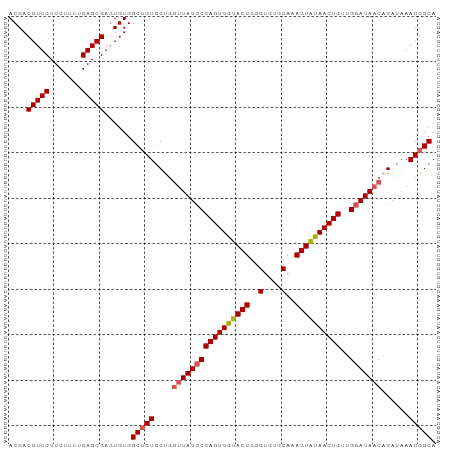
| Location | 14,045,808 – 14,045,902 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 95.96 |
| Shannon entropy | 0.07177 |
| G+C content | 0.46170 |
| Mean single sequence MFE | -33.16 |
| Consensus MFE | -30.24 |
| Energy contribution | -30.64 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.91 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.17 |
| SVM RNA-class probability | 0.984633 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 14045808 94 - 23011544 ACGACGUUCUUCUUUUGAGCGAGUGUGGCAGUGCUGGGUAUCCCAGUUGUACUGGGUGUGCAAAGUAUAACUGUUGUAUAAACGAUAAACGGCA .((.(((((.......))))).(((..((((((((..(((((((((.....))))).))))..)))...)))))..)))..........))... ( -31.20, z-score = -2.10, R) >droSim1.chr2L 13802832 94 - 22036055 ACGACGUUCUUCUUUUGAGCGAGUGUGGCUGUGCUGGUUAUCCCAGUUGUACUGGGUGUGCAAAGUAUAACUGUUGGAUAACAGAUAAACGGCA ....(((((.......)))))......(((((.(((.((((((((((((((((..(....)..))))))))))..)))))))))....))))). ( -35.30, z-score = -3.53, R) >droSec1.super_3 304096 94 - 7220098 ACGACGUUCUUCUUUUGAGCGAGUGUGGCUGUGCUGGUUAUCCCAGUUAUACUGGGUGUGCAAAGUAUAACUGUUGGAUAACAGAUAAACGGCA ....(((((.......)))))......(((((.(((.((((((((((((((((..(....)..))))))))))..)))))))))....))))). ( -35.30, z-score = -3.77, R) >droYak2.chr2L 14139034 94 - 22324452 ACGACGUUCUUCUUUUGAGCGACUGUGGCUGUGCUGGUUAUCCCAGUUGUACUGGGUGUGCAAAGUGUAACUGUUGGAUAACAGAUAAACGGCA ....(((((.......)))))......(((((.(((.((((((((((((((((..(....)..))))))))))..)))))))))....))))). ( -32.30, z-score = -2.39, R) >droEre2.scaffold_4929 15217171 94 + 26641161 ACGACGUUCUUCUUUUGAGCGAGUGUGGCUGUGCCGGUUAUCCCAGUUGUACUGGGUGUGCAAAGUGUAACUGUUGGAUAACAGAUAAACGGCA ....(((((.......)))))......(((((....(((((((((((((((((..(....)..))))))))))..)))))))......))))). ( -31.70, z-score = -1.97, R) >consensus ACGACGUUCUUCUUUUGAGCGAGUGUGGCUGUGCUGGUUAUCCCAGUUGUACUGGGUGUGCAAAGUAUAACUGUUGGAUAACAGAUAAACGGCA ....(((((.......)))))......(((((....(((((((((((((((((..(....)..))))))))))..)))))))......))))). (-30.24 = -30.64 + 0.40)


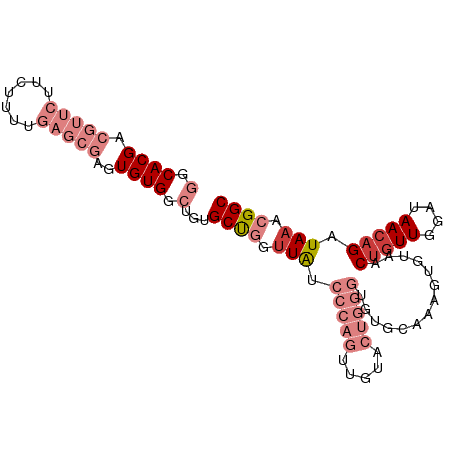
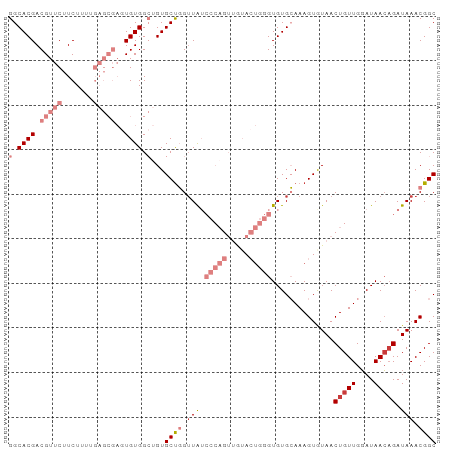
| Location | 14,045,809 – 14,045,905 |
|---|---|
| Length | 96 |
| Sequences | 7 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 82.24 |
| Shannon entropy | 0.32692 |
| G+C content | 0.48222 |
| Mean single sequence MFE | -31.70 |
| Consensus MFE | -12.08 |
| Energy contribution | -15.24 |
| Covariance contribution | 3.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.99 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.782204 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 14045809 96 - 23011544 GGCACGACGUUCUUCUUUUGAGCGAGUGUGGCAGUGCUGGGUAUCCCAGUUGUACUGGGUGUGCAAAGUAUAACUGUUGUAUAAACGAUAAACGGC .((.((.(((((.......))))).(((..((((((((..(((((((((.....))))).))))..)))...)))))..)))..........)))) ( -32.30, z-score = -2.19, R) >droSim1.chr2L 13802833 96 - 22036055 GGCACGACGUUCUUCUUUUGAGCGAGUGUGGCUGUGCUGGUUAUCCCAGUUGUACUGGGUGUGCAAAGUAUAACUGUUGGAUAACAGAUAAACGGC .((((..(((((.......))))).)))).(((((.(((.((((((((((((((((..(....)..))))))))))..)))))))))....))))) ( -38.20, z-score = -4.12, R) >droSec1.super_3 304097 96 - 7220098 GGCACGACGUUCUUCUUUUGAGCGAGUGUGGCUGUGCUGGUUAUCCCAGUUAUACUGGGUGUGCAAAGUAUAACUGUUGGAUAACAGAUAAACGGC .((((..(((((.......))))).)))).(((((.(((.((((((((((((((((..(....)..))))))))))..)))))))))....))))) ( -38.20, z-score = -4.41, R) >droYak2.chr2L 14139035 96 - 22324452 GGCACGACGUUCUUCUUUUGAGCGACUGUGGCUGUGCUGGUUAUCCCAGUUGUACUGGGUGUGCAAAGUGUAACUGUUGGAUAACAGAUAAACGGC ..((((.(((((.......)))))..))))(((((.(((.((((((((((((((((..(....)..))))))))))..)))))))))....))))) ( -33.20, z-score = -2.29, R) >droEre2.scaffold_4929 15217172 96 + 26641161 GGCACGACGUUCUUCUUUUGAGCGAGUGUGGCUGUGCCGGUUAUCCCAGUUGUACUGGGUGUGCAAAGUGUAACUGUUGGAUAACAGAUAAACGGC .((((..(((((.......))))).)))).(((((....(((((((((((((((((..(....)..))))))))))..)))))))......))))) ( -34.60, z-score = -2.45, R) >dp4.chr4_group4 967734 73 + 6586962 GGCACGACGGGCAUCUUU--------UGUGG--GUGCUUGUUG-------------GUGUGUGAAAAGUGUAACUGUUGGAUAACAGAUAAACGGC .(((((((((((((((..--------...))--))))))))).-------------)))).......((.((.(((((....))))).)).))... ( -22.70, z-score = -2.72, R) >droPer1.super_16 1330969 73 - 1990440 GGCACGACGGGCAUCUUU--------UGUGG--GUGCUUGUUG-------------GUGUGUGAAAAGUGUAACUGUUGGAUAACAGAUAAACGGC .(((((((((((((((..--------...))--))))))))).-------------)))).......((.((.(((((....))))).)).))... ( -22.70, z-score = -2.72, R) >consensus GGCACGACGUUCUUCUUUUGAGCGAGUGUGGCUGUGCUGGUUAUCCCAGUUGUACUGGGUGUGCAAAGUGUAACUGUUGGAUAACAGAUAAACGGC (.((((.(((((.......)))))..)))).)...((((.....(((((.....)))))..............(((((....))))).....)))) (-12.08 = -15.24 + 3.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:39:12 2011