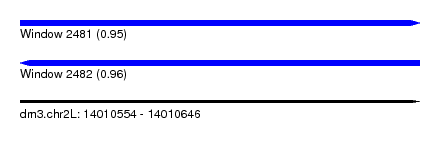
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 14,010,554 – 14,010,646 |
| Length | 92 |
| Max. P | 0.955972 |

| Location | 14,010,554 – 14,010,646 |
|---|---|
| Length | 92 |
| Sequences | 12 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 84.17 |
| Shannon entropy | 0.33138 |
| G+C content | 0.45240 |
| Mean single sequence MFE | -24.30 |
| Consensus MFE | -21.74 |
| Energy contribution | -21.67 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.89 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.951775 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
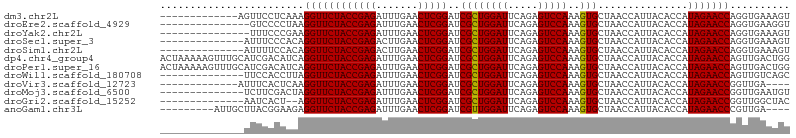
>dm3.chr2L 14010554 92 + 23011544 -------------AGUUCCUCAAAGGUUCUACCGAGAUUUGAACUCGGAUCGCUGGAUUCAGAGUCCAAAGUGCUAACCAUUACACCAUAGAACCAGGUGAAAGU -------------......(((..((((((((((((.......)))))..((((((((.....))))..))))...............)))))))...))).... ( -23.20, z-score = -1.21, R) >droEre2.scaffold_4929 15181501 90 - 26641161 ---------------GUCCCCUAAGGUUCUACCGAGAUUUGAACUCGGAUCGCUGGAUUCAGAGUCCAAAGUGCUAACCAUUACACCAUAGAACCAGGUGAAGGU ---------------.((.(((..((((((((((((.......)))))..((((((((.....))))..))))...............)))))))))).)).... ( -24.70, z-score = -1.39, R) >droYak2.chr2L 14102776 90 + 22324452 ---------------UUUCCCGAAGGUUCUACCGAGAUUUGAACUCGGAUCGCUGGAUUCAGAGUCCAAAGUGCUAACCAUUACACCAUAGAACCAGGUGAAAGU ---------------((((((...((((((((((((.......)))))..((((((((.....))))..))))...............))))))).)).)))).. ( -25.90, z-score = -2.35, R) >droSec1.super_3 267495 91 + 7220098 --------------AUUUCCCACAGGUUCUACCGAGACUUGAACUCGGAUCGCUGGAUUCAGAGUCCAAAGUGCUAACCAUUACACCAUAGAACCAGGUGAAAGU --------------.((((((...((((((((((((.......)))))..((((((((.....))))..))))...............))))))).)).)))).. ( -25.10, z-score = -2.10, R) >droSim1.chr2L 13766961 91 + 22036055 --------------AUUUUCCACAGGUUCUACCGAGACUUGAACUCGGAUCGCUGGAUUCAGAGUCCAAAGUGCUAACCAUUACACCAUAGAACCAGGUGAAAGU --------------((((((.((.((((((((((((.......)))))..((((((((.....))))..))))...............)))))))..)))))))) ( -24.80, z-score = -1.98, R) >dp4.chr4_group4 913088 105 - 6586962 ACUAAAAAGUUUGCAUCGACAUCAGGUUCUACCGAGAUUUGAACUCGGAUCGCUGGAUUCAGAGUCCAAAGUGCUAACCAUUACACCAUAGAACCAGUUGACUGG .............(((((((....((((((((((((.......)))))..((((((((.....))))..))))...............))))))).))))).)). ( -25.10, z-score = -1.17, R) >droPer1.super_16 1273088 105 + 1990440 ACUAAAAAGUUUGCAUCGACAUCAGGUUCUACCGAGAUUUGAACUCGGAUCGCUGGAUUCAGAGUCCAAAGUGCUAACCAUUACACCAUAGAACCAGUUGACUGG .............(((((((....((((((((((((.......)))))..((((((((.....))))..))))...............))))))).))))).)). ( -25.10, z-score = -1.17, R) >droWil1.scaffold_180708 11034907 91 - 12563649 --------------UUCCACCUUAGGUUCUACCGAGAUUUGAACUCGGAUCGCUGGAUUCAGAGUCCAAAGUGCUAACCAUUACACCAUAGAACCAGUUGUCAGC --------------..........((((((((((((.......)))))..((((((((.....))))..))))...............))))))).......... ( -22.10, z-score = -1.56, R) >droVir3.scaffold_12723 1062357 88 + 5802038 -------------AUUUCACUCAAGGUUCUACCGAGAUUUGAACUCGGAUCGCUGGAUUCAGAGUCCAAAGUGCUAACCAUUACACCAUAGAACCGGUUGA---- -------------...........((((((((((((.......)))))..((((((((.....))))..))))...............)))))))......---- ( -22.10, z-score = -1.28, R) >droMoj3.scaffold_6500 22042048 91 - 32352404 --------------UCUUCGACUAGGUUCUACCGAGAUUUGAACUCGGAUCGCUGGAUUCAGAGUCCAAAGUGCUAACCAUUACACCAUAGAACCGGUUGAAUGU --------------..(((((((.((((((((((((.......)))))..((((((((.....))))..))))...............))))))))))))))... ( -28.30, z-score = -2.71, R) >droGri2.scaffold_15252 4508651 89 + 17193109 --------------AAUCACU--AGGUUCUACCGAGAUUUGAACUCGGAUCGCUGGAUUCAGAGUCCAAAGUGCUAACCAUUACACCAUAGAACCGGUUGGCUAC --------------..(((..--.((((((((((((.......)))))..((((((((.....))))..))))...............)))))))...))).... ( -22.50, z-score = -1.26, R) >anoGam1.chr3L 13298801 92 - 41284009 ---------AUUGCUUACGGAAGAGGUUCUACCGAGAUUUGAACUCGGAUCGUUGGAUUCAGAGUCCAAAGUGCUAACCAUUACACCAUAGAACCCGUUGA---- ---------........(....).((((((((((((.......)))))....((((((.....))))))...................)))))))......---- ( -22.70, z-score = -0.99, R) >consensus ______________AUUCACCAAAGGUUCUACCGAGAUUUGAACUCGGAUCGCUGGAUUCAGAGUCCAAAGUGCUAACCAUUACACCAUAGAACCAGUUGAAAGU ........................((((((((((((.......)))))..((((((((.....)))))..)))...............))))))).......... (-21.74 = -21.67 + -0.08)
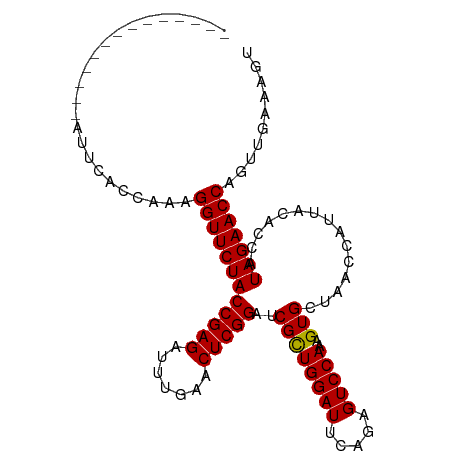
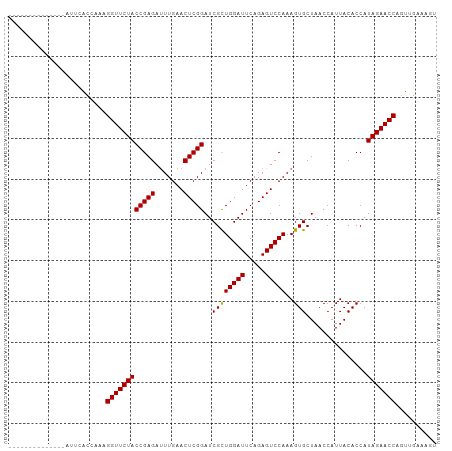
| Location | 14,010,554 – 14,010,646 |
|---|---|
| Length | 92 |
| Sequences | 12 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 84.17 |
| Shannon entropy | 0.33138 |
| G+C content | 0.45240 |
| Mean single sequence MFE | -26.96 |
| Consensus MFE | -24.48 |
| Energy contribution | -24.48 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.91 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.955972 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 14010554 92 - 23011544 ACUUUCACCUGGUUCUAUGGUGUAAUGGUUAGCACUUUGGACUCUGAAUCCAGCGAUCCGAGUUCAAAUCUCGGUAGAACCUUUGAGGAACU------------- ..(((((...(((((((((((((........)))))((((((((.((.((....)))).))))))))......))))))))..)))))....------------- ( -26.90, z-score = -1.55, R) >droEre2.scaffold_4929 15181501 90 + 26641161 ACCUUCACCUGGUUCUAUGGUGUAAUGGUUAGCACUUUGGACUCUGAAUCCAGCGAUCCGAGUUCAAAUCUCGGUAGAACCUUAGGGGAC--------------- ...(((.((((((((((((((((........)))))((((((((.((.((....)))).))))))))......))))))))..)))))).--------------- ( -27.70, z-score = -1.35, R) >droYak2.chr2L 14102776 90 - 22324452 ACUUUCACCUGGUUCUAUGGUGUAAUGGUUAGCACUUUGGACUCUGAAUCCAGCGAUCCGAGUUCAAAUCUCGGUAGAACCUUCGGGAAA--------------- ...(((.((.(((((((((((((........)))))((((((((.((.((....)))).))))))))......))))))))...))))).--------------- ( -27.80, z-score = -1.93, R) >droSec1.super_3 267495 91 - 7220098 ACUUUCACCUGGUUCUAUGGUGUAAUGGUUAGCACUUUGGACUCUGAAUCCAGCGAUCCGAGUUCAAGUCUCGGUAGAACCUGUGGGAAAU-------------- ..(..(((..(((((((((((((........)))))((((((((.((.((....)))).))))))))......)))))))).)))..)...-------------- ( -30.10, z-score = -2.28, R) >droSim1.chr2L 13766961 91 - 22036055 ACUUUCACCUGGUUCUAUGGUGUAAUGGUUAGCACUUUGGACUCUGAAUCCAGCGAUCCGAGUUCAAGUCUCGGUAGAACCUGUGGAAAAU-------------- ..((((((..(((((((((((((........)))))((((((((.((.((....)))).))))))))......)))))))).))))))...-------------- ( -30.30, z-score = -2.71, R) >dp4.chr4_group4 913088 105 + 6586962 CCAGUCAACUGGUUCUAUGGUGUAAUGGUUAGCACUUUGGACUCUGAAUCCAGCGAUCCGAGUUCAAAUCUCGGUAGAACCUGAUGUCGAUGCAAACUUUUUAGU ...(((.((.(((((((((((((........)))))((((((((.((.((....)))).))))))))......))))))))....)).))).............. ( -26.70, z-score = -0.75, R) >droPer1.super_16 1273088 105 - 1990440 CCAGUCAACUGGUUCUAUGGUGUAAUGGUUAGCACUUUGGACUCUGAAUCCAGCGAUCCGAGUUCAAAUCUCGGUAGAACCUGAUGUCGAUGCAAACUUUUUAGU ...(((.((.(((((((((((((........)))))((((((((.((.((....)))).))))))))......))))))))....)).))).............. ( -26.70, z-score = -0.75, R) >droWil1.scaffold_180708 11034907 91 + 12563649 GCUGACAACUGGUUCUAUGGUGUAAUGGUUAGCACUUUGGACUCUGAAUCCAGCGAUCCGAGUUCAAAUCUCGGUAGAACCUAAGGUGGAA-------------- ..(.((....(((((((((((((........)))))((((((((.((.((....)))).))))))))......))))))))....)).)..-------------- ( -25.70, z-score = -1.10, R) >droVir3.scaffold_12723 1062357 88 - 5802038 ----UCAACCGGUUCUAUGGUGUAAUGGUUAGCACUUUGGACUCUGAAUCCAGCGAUCCGAGUUCAAAUCUCGGUAGAACCUUGAGUGAAAU------------- ----((((..(((((((((((((........)))))((((((((.((.((....)))).))))))))......)))))))))))).......------------- ( -25.60, z-score = -1.85, R) >droMoj3.scaffold_6500 22042048 91 + 32352404 ACAUUCAACCGGUUCUAUGGUGUAAUGGUUAGCACUUUGGACUCUGAAUCCAGCGAUCCGAGUUCAAAUCUCGGUAGAACCUAGUCGAAGA-------------- ...(((.((.(((((((((((((........)))))((((((((.((.((....)))).))))))))......))))))))..)).)))..-------------- ( -25.70, z-score = -1.40, R) >droGri2.scaffold_15252 4508651 89 - 17193109 GUAGCCAACCGGUUCUAUGGUGUAAUGGUUAGCACUUUGGACUCUGAAUCCAGCGAUCCGAGUUCAAAUCUCGGUAGAACCU--AGUGAUU-------------- .....((...(((((((((((((........)))))((((((((.((.((....)))).))))))))......)))))))).--..))...-------------- ( -25.10, z-score = -1.44, R) >anoGam1.chr3L 13298801 92 + 41284009 ----UCAACGGGUUCUAUGGUGUAAUGGUUAGCACUUUGGACUCUGAAUCCAACGAUCCGAGUUCAAAUCUCGGUAGAACCUCUUCCGUAAGCAAU--------- ----.....((((((((((((((........)))))((((((((.((.((....)))).))))))))......)))))))))..............--------- ( -25.20, z-score = -1.70, R) >consensus ACUUUCAACUGGUUCUAUGGUGUAAUGGUUAGCACUUUGGACUCUGAAUCCAGCGAUCCGAGUUCAAAUCUCGGUAGAACCUGAGGGGAAU______________ ..........(((((((((((((........)))))((((((((.((.((....)))).))))))))......))))))))........................ (-24.48 = -24.48 + -0.00)

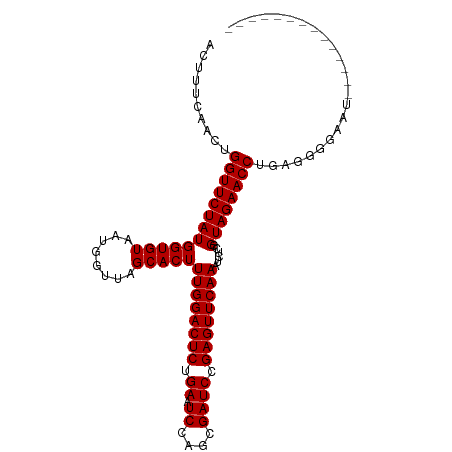
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:39:09 2011