
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 13,997,786 – 13,997,879 |
| Length | 93 |
| Max. P | 0.756537 |

| Location | 13,997,786 – 13,997,879 |
|---|---|
| Length | 93 |
| Sequences | 9 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 80.61 |
| Shannon entropy | 0.39222 |
| G+C content | 0.42863 |
| Mean single sequence MFE | -23.60 |
| Consensus MFE | -17.43 |
| Energy contribution | -18.22 |
| Covariance contribution | 0.79 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.756537 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 13997786 93 - 23011544 GGACACAUUUUCAGGUCAGUUGCCAGUUGGCAGCUGCCUUUGUGCUCGAUUGUGCUCUAUUGAUUGCCAGCCAAGAGAACCAAAAAAAUGAUA--- ((.((((.....(((.((((((((....))))))))))).)))).((..(((.(((............))))))..)).))............--- ( -26.30, z-score = -0.96, R) >droSim1.chr2L 13754071 92 - 22036055 GGACACAUUUUCAGGUCAGUUGCCAGUUGGCAGCUGCCUUUGUGCUCGAUUGUGCUCUAUUGAUUGCCAGCCAAGAGAACC-AAAAAAUGAUA--- ((.((((.....(((.((((((((....))))))))))).)))).((..(((.(((............))))))..)).))-...........--- ( -26.30, z-score = -0.94, R) >droSec1.super_3 254730 92 - 7220098 GGACACAUUUUCAGGUCAGUUGCCAGUUGGCAGCUGCCUUUGUGCUCGAUUGUGCUCUAUUGAUUGCCAGCCAAGAGAACC-AAAAAAUGAUA--- ((.((((.....(((.((((((((....))))))))))).)))).((..(((.(((............))))))..)).))-...........--- ( -26.30, z-score = -0.94, R) >droYak2.chr2L 14089725 89 - 22324452 GGACACAUUUUCAGGUCAGUUGCCAGUUGGCAGCUGCCUUUGUGCUCGAU---ACUCUAUUGAUUGCCAGCCAAGAGAACC-AAAAAAUGAUA--- (..((((.....(((.((((((((....))))))))))).))))..)...---.((((.(((.....)))...))))....-...........--- ( -24.90, z-score = -1.37, R) >droEre2.scaffold_4929 15168727 89 + 26641161 GGACACAUUUUCAGGUCAGUUGCCAGUUGGCAGCUGCCUUUGUGCUCGAU---ACUCUAUUGAUUGCCAGCCAAGAGAACC-AAAAAAUGAUA--- (..((((.....(((.((((((((....))))))))))).))))..)...---.((((.(((.....)))...))))....-...........--- ( -24.90, z-score = -1.37, R) >droAna3.scaffold_12916 4397227 92 + 16180835 GCACACAUUUUCAGGUCAGUUGCCAGUUGGCAGCCGCCUUUGUGCUCGAC---AUUCUAUUGAUUGCCAGCCAAGAACAAA-AAAGAGUACUCAUA ............((((..((((((....)))))).))))..((((((...---.((((.(((.....)))...))))....-...))))))..... ( -20.50, z-score = -0.23, R) >dp4.chr4_group4 895615 85 + 6586962 GAACACAUUUUCAGGUCAGUUGCCAGUUGGCAGCUGCCUUUGUGCGCGAU---GUUCUAUUGAUUUCCAUACAAAUAAAU--AUCUUGUA------ (..((((.....(((.((((((((....))))))))))).))))..)(((---(((.((((............)))))))--))).....------ ( -24.20, z-score = -2.63, R) >droPer1.super_16 1255416 85 - 1990440 GAACACAUUUUCAGGUCAGUUGCCAGUUGGCAGCUGCCUUUGUGCGCGAU---GUUCUAUUGAUUUCCAUACAAAAAAAU--AUAUUGUA------ ...((((.....(((.((((((((....))))))))))).)))).(((((---((....(((.........)))......--))))))).------ ( -22.60, z-score = -2.02, R) >droWil1.scaffold_180708 11012667 83 + 12563649 GGAAACAUUUUCAGGUCACUUGCCAGUUGGCAGCUCUCUAG------------GUUCUAUUGAUUGCCAAAAAAAAAAAAGAAAAAAGAAGAAUA- (....).(((((.(((.....)))..(((((((.((..(((------------...)))..))))))))).................)))))...- ( -16.40, z-score = -0.73, R) >consensus GGACACAUUUUCAGGUCAGUUGCCAGUUGGCAGCUGCCUUUGUGCUCGAU___GCUCUAUUGAUUGCCAGCCAAGAGAACC_AAAAAAUGAUA___ (..((((.....(((.((((((((....))))))))))).))))..)................................................. (-17.43 = -18.22 + 0.79)

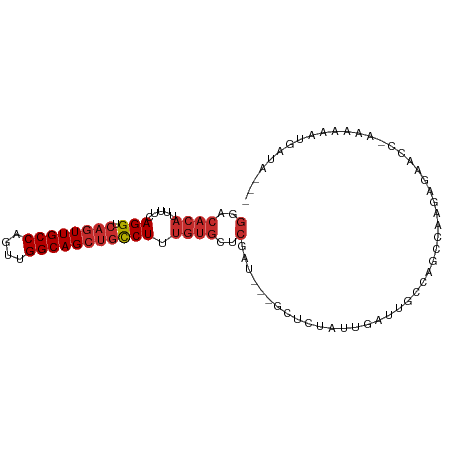
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:39:07 2011