| Sequence ID | dm3.chr2L |
|---|---|
| Location | 13,973,822 – 13,973,924 |
| Length | 102 |
| Max. P | 0.999027 |

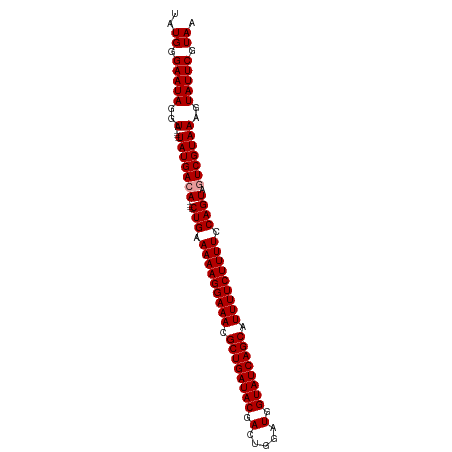
| Location | 13,973,822 – 13,973,924 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 91.28 |
| Shannon entropy | 0.11704 |
| G+C content | 0.36914 |
| Mean single sequence MFE | -37.23 |
| Consensus MFE | -27.37 |
| Energy contribution | -27.70 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -6.46 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.60 |
| SVM RNA-class probability | 0.999027 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 13973822 102 + 23011544 UAUGGGAAUAGAUACAUAUAUGACAAGUACUGAAAAAGGAAACGCUGAUACGACUGGUUGGUAUCAGCGUUUUCUUUUCCAGUACUCGUAAAGUAUUCGUAU ...........((((..((((.((.(((((((.((((((((((((((((((.(.....).)))))))))))))))))).))))))).))...))))..)))) ( -41.10, z-score = -7.20, R) >droSim1.chr2L 13729867 94 + 22036055 UAUGGGAAUAGGUA----UAUGACA----CUGAAAAAGGAAACGCUGAUACGACUGGAUGGUAUCAGCAUUUUCUUUUCCAGUAGUCGUAAAGUAUUCGUAA ..((.(((((..(.----(((((((----(((.(((((((((.((((((((.(.....).)))))))).))))))))).)))).)))))))..))))).)). ( -35.30, z-score = -6.09, R) >droSec1.super_3 230568 94 + 7220098 UAUGGGAAUAGGUA----UAUGACA----CUGAAAAAGGAAACGCUGAUACGACUGGAUGGUAUCAGCAUUUUCUUUUCCAGUAGUCGUAAAGUAUUCGUAA ..((.(((((..(.----(((((((----(((.(((((((((.((((((((.(.....).)))))))).))))))))).)))).)))))))..))))).)). ( -35.30, z-score = -6.09, R) >consensus UAUGGGAAUAGGUA____UAUGACA____CUGAAAAAGGAAACGCUGAUACGACUGGAUGGUAUCAGCAUUUUCUUUUCCAGUAGUCGUAAAGUAUUCGUAA ..((.(((((........((((((.....(((.(((((((((.((((((((.(.....).)))))))).))))))))).)))..))))))...))))).)). (-27.37 = -27.70 + 0.33)

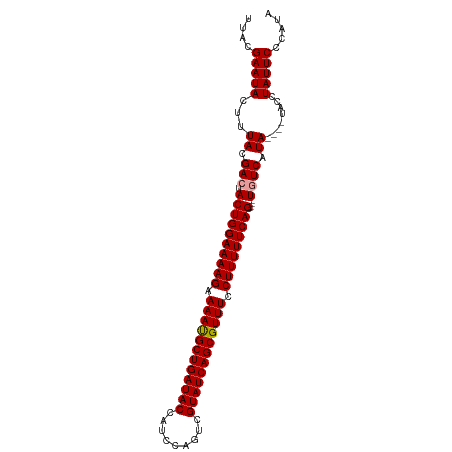
| Location | 13,973,822 – 13,973,924 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 91.28 |
| Shannon entropy | 0.11704 |
| G+C content | 0.36914 |
| Mean single sequence MFE | -35.67 |
| Consensus MFE | -27.45 |
| Energy contribution | -27.57 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -8.43 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.50 |
| SVM RNA-class probability | 0.991873 |
| Prediction | RNA |
| WARNING | Mean z-score out of range. |
Download alignment: ClustalW | MAF
>dm3.chr2L 13973822 102 - 23011544 AUACGAAUACUUUACGAGUACUGGAAAAGAAAACGCUGAUACCAACCAGUCGUAUCAGCGUUUCCUUUUUCAGUACUUGUCAUAUAUGUAUCUAUUCCCAUA (((((.(((....((((((((((((((((.((((((((((((.........)))))))))))).))))))))))))))))..))).)))))........... ( -40.80, z-score = -9.26, R) >droSim1.chr2L 13729867 94 - 22036055 UUACGAAUACUUUACGACUACUGGAAAAGAAAAUGCUGAUACCAUCCAGUCGUAUCAGCGUUUCCUUUUUCAG----UGUCAUA----UACCUAUUCCCAUA ....(((((...((.(((.((((((((((.((((((((((((.........)))))))))))).)))))))))----)))).))----....)))))..... ( -33.10, z-score = -8.01, R) >droSec1.super_3 230568 94 - 7220098 UUACGAAUACUUUACGACUACUGGAAAAGAAAAUGCUGAUACCAUCCAGUCGUAUCAGCGUUUCCUUUUUCAG----UGUCAUA----UACCUAUUCCCAUA ....(((((...((.(((.((((((((((.((((((((((((.........)))))))))))).)))))))))----)))).))----....)))))..... ( -33.10, z-score = -8.01, R) >consensus UUACGAAUACUUUACGACUACUGGAAAAGAAAAUGCUGAUACCAUCCAGUCGUAUCAGCGUUUCCUUUUUCAG____UGUCAUA____UACCUAUUCCCAUA ...............(((..(((((((((.((((((((((((.........)))))))))))).))))))))).....)))..................... (-27.45 = -27.57 + 0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:39:05 2011