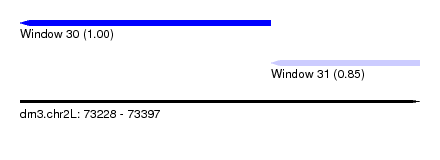
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 73,228 – 73,397 |
| Length | 169 |
| Max. P | 0.995075 |

| Location | 73,228 – 73,334 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 64.74 |
| Shannon entropy | 0.61111 |
| G+C content | 0.36808 |
| Mean single sequence MFE | -26.84 |
| Consensus MFE | -9.52 |
| Energy contribution | -10.64 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.37 |
| Mean z-score | -2.97 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.76 |
| SVM RNA-class probability | 0.995075 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
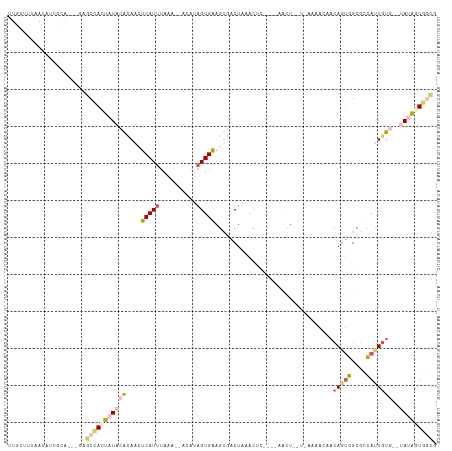
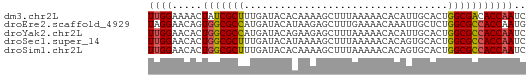
>dm3.chr2L 73228 106 - 23011544 CUGCUUUAAAAUCGCAGUGGCGCCAUUAUACACAACUCAUUUAAACUACAUAGUGAAGUGCCUGAACUU----AA-U--U-AAAACAACAGUGGCGCCAUUGUG--UAUAUUGGCG ..(((.......(((((((((((((((...(((...................)))((((......))))----..-.--.-........)))))))))))))))--......))). ( -32.53, z-score = -3.65, R) >droSim1.chr2L 73306 102 - 22036055 UUGGUUUAAUAUCGCA---GAGCCACUAUACACAACUCAUUUAAA--ACAUAGUGAAGUGACUAAACUC----AACU--U-AAAACUACAGUGGCGCCAUUGUG--UAUAGUGGCG ................---..((((((((((((............--...((((.((((((......))----.)))--)-...)))).((((....)))))))--))))))))). ( -28.90, z-score = -3.30, R) >droSec1.super_14 72364 102 - 2068291 UUGGUUUAAUAUCGCA---GAGCCACUAUACACAACUCAUUUAAA--ACAUAGUGAAGUGACUAAACUC----AACU--U-AAAACUACAGUGGCGCCAUUGUG--UAUAGUGGCG ................---..((((((((((((............--...((((.((((((......))----.)))--)-...)))).((((....)))))))--))))))))). ( -28.90, z-score = -3.30, R) >droYak2.chr2L 62571 112 - 22324452 AUGCUUUAAUAAAACAGUGACACCACUAUACAAAAAUCAUUUAAA--CAUUAGUGAAGCCACUAAACCCAUCUAAUU--UAAAAAAAACAGUGGCGGCACUAUACUCACAUUAAAG ...(((((((.....((((....))))..................--...(((((..((((((..............--..........))))))..))))).......))))))) ( -20.56, z-score = -3.89, R) >droGri2.scaffold_15252 17107988 109 - 17193109 GUGCGUAAAACUUGA----AUGCUAGGUUAAGUAUUACAUAUAAUAUACAGGGUGUAGCG-CUACAGCAAAACAGCUGAUGAAAAGACCACCAGAAUUAUUGUA--CAUUCUGAUG ((((((((.((((((----........)))))).))))......(((((...))))))))-)..((((......)))).............((((((.......--.))))))... ( -23.30, z-score = -0.73, R) >consensus UUGCUUUAAUAUCGCA___GAGCCACUAUACACAACUCAUUUAAA__ACAUAGUGAAGUGACUAAACUC____AACU__U_AAAACAACAGUGGCGCCAUUGUG__UAUAGUGGCG .....................((((((((.......(((((..........)))))...............................((((((....))))))....)))))))). ( -9.52 = -10.64 + 1.12)

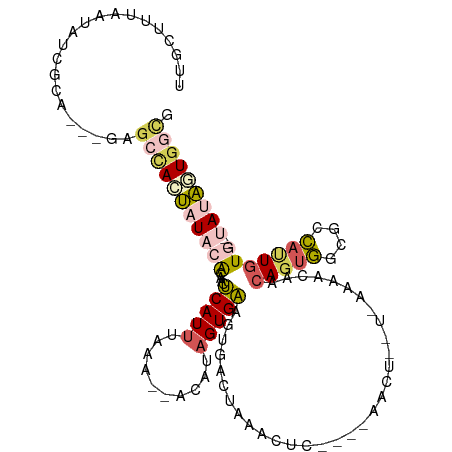
| Location | 73,334 – 73,397 |
|---|---|
| Length | 63 |
| Sequences | 5 |
| Columns | 63 |
| Reading direction | reverse |
| Mean pairwise identity | 88.57 |
| Shannon entropy | 0.20040 |
| G+C content | 0.44444 |
| Mean single sequence MFE | -15.77 |
| Consensus MFE | -10.74 |
| Energy contribution | -10.58 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.849417 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
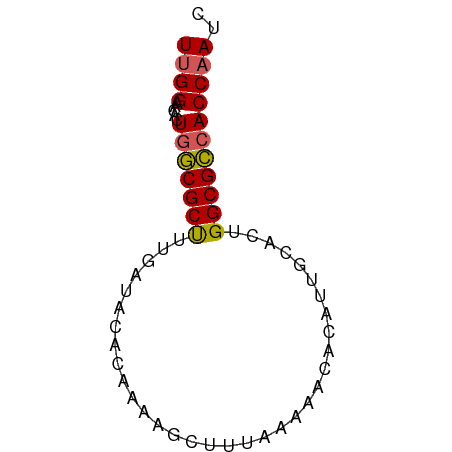
>dm3.chr2L 73334 63 - 23011544 UUGGAAAACUAUCGCUUUGAUACACAAAAGCUUUAAAAACACAUUGCACUGGCGACACCAAUC ((((.........(((((........)))))............((((....))))..)))).. ( -10.20, z-score = -2.28, R) >droEre2.scaffold_4929 100584 63 - 26641161 UAGGAACAGUGGCGCCAUGAUACAUAAGAGCUUUGAAAACAAAUUGCUCUGGCGCCACCAAUG ........(((((((((((...))).(((((((((....))))..)))))))))))))..... ( -22.80, z-score = -4.37, R) >droYak2.chr2L 62798 63 - 22324452 UUGGAACACUGGCGCCAUGAUACAGAAGAGCUUUAAAAACACAUUGCACUGGCGCCACCAAUC ((((.....((((((((.....(....).((..............))..)))))))))))).. ( -15.44, z-score = -2.14, R) >droSec1.super_14 72691 63 - 2068291 UUGGAACACUGGCGCUUUGAUACAUAAAAGCUUUAAAAACACAGUGCACUGGCGCCACCAAUC ((((.....(((((((((((..(......)..)))))....(((....))))))))))))).. ( -15.20, z-score = -1.88, R) >droSim1.chr2L 73633 63 - 22036055 UUGGAACACUGGCGCUUUGAUACACAAAAGCUUUAAAAACACAGUGCACUGGCGCCACCAAUC ((((.....((((((((((.....)))).............(((....))))))))))))).. ( -15.20, z-score = -1.77, R) >consensus UUGGAACACUGGCGCUUUGAUACACAAAAGCUUUAAAAACACAUUGCACUGGCGCCACCAAUC ((((.....(((((((..................................))))))))))).. (-10.74 = -10.58 + -0.16)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:05:05 2011