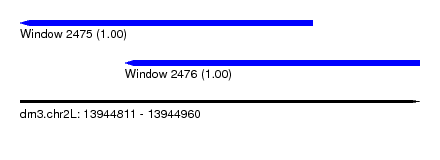
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 13,944,811 – 13,944,960 |
| Length | 149 |
| Max. P | 0.999533 |

| Location | 13,944,811 – 13,944,920 |
|---|---|
| Length | 109 |
| Sequences | 11 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 76.26 |
| Shannon entropy | 0.49738 |
| G+C content | 0.34074 |
| Mean single sequence MFE | -28.45 |
| Consensus MFE | -20.19 |
| Energy contribution | -19.31 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.55 |
| Mean z-score | -2.93 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.76 |
| SVM RNA-class probability | 0.999285 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
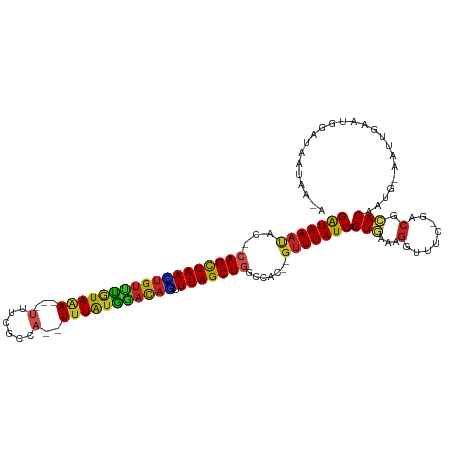
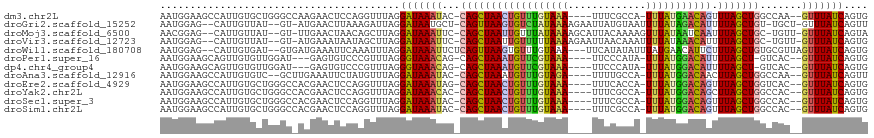
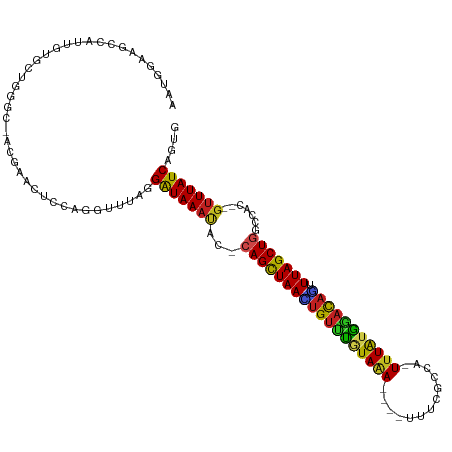
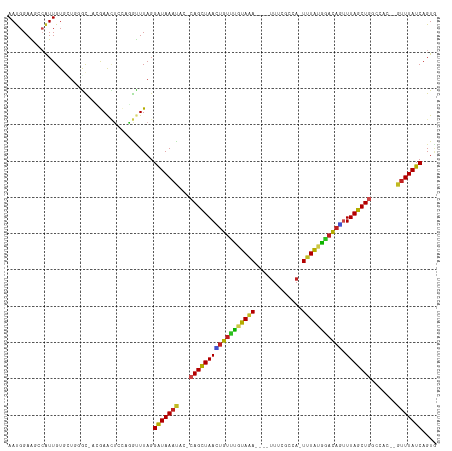
>dm3.chr2L 13944811 109 - 23011544 GAUAAAUAC-CAGCUAACUGUUUGUAAA---UUUCGCCA--UUUAUGAACAGUUUAGCUGGCCAA--GUUUAUCAGUGAAAGGUUUC-GGCGCACAAUG-AAUUGAAUGGAUAAUAAGA (((((((.(-((((((((((((..((((---(......)--))))..))))).))))))))....--))))))).(((....((...-.)).)))....-................... ( -29.50, z-score = -2.84, R) >droGri2.scaffold_15252 4431505 114 - 17193109 GAUAAUGCU-CAGUUAAGUGUCUAUAAAAGAAUUAUGUAAUUUUAUAGACAUUUUAGCUGU-UGCU-GUUUAUCAGUUAAAGGUUUC-AACGUACAAUG-AAUGAAAAAUGUAUAAAAA (((((.((.-(((((((((((((((((((...........))))))))))).)))))))).-.)).-..))))).............-...(((((.(.-.......).)))))..... ( -26.60, z-score = -3.26, R) >droMoj3.scaffold_6500 21944341 111 + 32352404 GAUAAAUUC-CAGCUAAUUGUUUAUAAAAGCAUUACAAAAGUUUAUAAUCAAUUUAGCUGC-UGUU-GUUUAUCAGUAGAAGGUUUC-GACGUACAAUGAAAUGAUAUAUGUAAA---- (((((((..-((((.(((((.(((((((.............))))))).))))).....))-))..-)))))))........(((((-(........))))))............---- ( -19.72, z-score = -0.93, R) >droWil1.scaffold_180708 10935779 105 + 12563649 GAUAAAUUCUCAGUUAAGUGUUUGUAAA---UUCAUAUAUUUAUGAACAUUCUUUAGCUGUGCGUUAGUUUAUCAGUGAAAGGUUUCAAAAGCACAAUG-AACAACAAA---------- (((((((((.((((((((((((..((((---(......)))))..)))))...))))))).)....)))))))).(((..............)))....-.........---------- ( -21.14, z-score = -1.69, R) >droPer1.super_16 1192876 106 - 1990440 GGUAAACAG-CAGCUAAAUGUUCGUAAA---UUCCCAUA--UUUAUGGACAUUUUAGCU-GUCAC--GUUUAUCAGUGAAAGGUUUC-AACGCACAAUG-AAUUGAAUAGACAGCUA-- (((((((.(-((((((((((((((((((---(......)--)))))))))).)))))))-))...--)))))))(((.......(((-((..(.....)-..)))))......))).-- ( -32.82, z-score = -4.73, R) >dp4.chr4_group4 838206 106 + 6586962 GGUAAACAG-CAGCUAAAUGUUCGUAAA---UUCCCAUA--UUUAUGGACAUUUUAGCU-GUCAC--GUUUAUCAGUGAAAGGUUUC-AACGCACAAUG-AAUUGAAUAGACAGCUA-- (((((((.(-((((((((((((((((((---(......)--)))))))))).)))))))-))...--)))))))(((.......(((-((..(.....)-..)))))......))).-- ( -32.82, z-score = -4.73, R) >droAna3.scaffold_12916 4346712 107 + 16180835 GAUAAAUAC-CAGCUAAAUGUUUGUAGA---UUUUGCCA--UUUAUGGACAACUUAGCUGGCCAA--GUUUAUCAGUUAAAGGUUUC-ACCGCACAAUG-AAUUGAAUGGAUAACCA-- (((((((.(-(((((((.((((..((((---(......)--))))..))))..))))))))....--))))))).......((((..-.(((..(((..-..)))..)))..)))).-- ( -30.80, z-score = -3.67, R) >droEre2.scaffold_4929 15114228 108 + 26641161 GAUAAAUAG-CAGCUAACUGUUUGUAAA---UUUCACCA--UUUAUGGACAGUUUAGCUGGUCAC--GUUUAUCAGUG-AAGGUUUC-GGCGCACAAUG-AAUUGAAUGGAUAAUAAGA .........-((((((((((((..((((---(......)--))))..))))).)))))))(((.(--(((((((((((-...((...-.)).)))..))-)..)))))))))....... ( -28.40, z-score = -2.48, R) >droYak2.chr2L 14028939 109 - 22324452 GAUAAACAC-CAGCUAACUGUUUGUAAA---UUUCGCCA--UUUAUGGACAGCUUAGCUGGCCAC--GUUUAUCAGUGAAAGGUUUC-GGCGCACAAUG-AAUUGAAUGGAUAAUAAGA (((((((.(-((((((((((((..((((---(......)--))))..))))).))))))))....--))))))).(((....((...-.)).)))....-................... ( -31.40, z-score = -2.83, R) >droSec1.super_3 202027 109 - 7220098 GAUAAAUAC-CAGCUAACUGUUUGUAAA---UUUCGCCA--UUUAUGGACAGUUUAGCUGGCCAC--GUUUAUCAGUGAAAGGUUUC-GGCGCACAAUG-AAUUGAAUGGAUAAUAAGA (((((((.(-((((((((((((..((((---(......)--))))..))))).))))))))....--))))))).(((....((...-.)).)))....-................... ( -29.90, z-score = -2.56, R) >droSim1.chr2L 13699010 109 - 22036055 GAUAAAUAC-CAGCUAACUGUUUGUAAA---UUUCGCCA--UUUAUGGACAGUUUAGCUGGCCAC--GUUUAUCAGUGAAAGGUUUC-GGCGCACAAUG-AAUUGAAUGGAUAAUAAGA (((((((.(-((((((((((((..((((---(......)--))))..))))).))))))))....--))))))).(((....((...-.)).)))....-................... ( -29.90, z-score = -2.56, R) >consensus GAUAAAUAC_CAGCUAACUGUUUGUAAA___UUUCGCCA__UUUAUGGACAGUUUAGCUGGCCAC__GUUUAUCAGUGAAAGGUUUC_GACGCACAAUG_AAUUGAAUGGAUAAUAA_A (((((((...((((((((((((((((((.............))))))))))).))))))).......))))))).(((...(........).)))........................ (-20.19 = -19.31 + -0.88)
| Location | 13,944,850 – 13,944,960 |
|---|---|
| Length | 110 |
| Sequences | 12 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 73.86 |
| Shannon entropy | 0.55332 |
| G+C content | 0.36711 |
| Mean single sequence MFE | -32.23 |
| Consensus MFE | -19.21 |
| Energy contribution | -18.39 |
| Covariance contribution | -0.82 |
| Combinations/Pair | 1.60 |
| Mean z-score | -2.97 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.98 |
| SVM RNA-class probability | 0.999533 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2L 13944850 110 - 23011544 AAUGGAAGCCAUUGUGCUGGGCCAAGAACUCCAGGUUUAGGAUAAAUAC-CAGCUAACUGUUUGUAAA----UUUCGCCA-UUUAUGAACAGUUUAGCUGGCCAA--GUUUAUCAGUG (((((...)))))...(((((((..(....)..)))))))(((((((.(-((((((((((((..((((----(......)-))))..))))).))))))))....--))))))).... ( -36.60, z-score = -3.37, R) >droGri2.scaffold_15252 4431544 110 - 17193109 AAUGGAG--CAUUGUUAU--GU-AUGAACUUAAAGAUUAGGAUAAUGCU-CAGUUAAGUGUCUAUAAAAGAAUUAUGUAAUUUUAUAGACAUUUUAGCUGU-UGCU-GUUUAUCAGUU ....(((--(((((((..--..-.(((.(.....).))).)))))))))-)((((((((((((((((((...........))))))))))).)))))))..-.(((-(.....)))). ( -29.70, z-score = -3.49, R) >droMoj3.scaffold_6500 21944377 110 + 32352404 AACGGAG--CAUUGUUAU--GU-UUGAACUAACAGCUUAGGAUAAAUUC-CAGCUAAUUGUUUAUAAAAGCAUUACAAAAGUUUAUAAUCAAUUUAGCUGC-UGUU-GUUUAUCAGUA ....(((--(((....))--))-))((((.((((((...(((.....))-)(((((((((.(((((((.............))))))).))).))))))))-))))-))))....... ( -26.42, z-score = -2.26, R) >droVir3.scaffold_12723 926431 110 - 5802038 AAUGGAG--CAUUGUUAU--GU-AUGAAAUAAUAGCUUAGGAUAAAUUC-CAGCUAAUUGUUUUUAAAAGAAUUAACAAAUUUUAUAAACAUUUUAGCUGC-UGUU-GUUUAUCAGUG ..(((.(--(((....))--))-...((((((((((...(((.....))-)((((((.(((((.(((((...........))))).)))))..))))))))-))))-)))).)))... ( -25.60, z-score = -2.57, R) >droWil1.scaffold_180708 10935809 111 + 12563649 AAUGGAG--CAUUGUGAU--GUGAUGAAAUUCAAAUUUAGGAUAAAUUCUCAGUUAAGUGUUUGUAAA---UUCAUAUAUUUAUGAACAUUCUUUAGCUGUGCGUUAGUUUAUCAGUG ..((((.--(((..(...--)..)))...)))).......(((((((((.((((((((((((..((((---(......)))))..)))))...))))))).)....)))))))).... ( -24.30, z-score = -1.73, R) >droPer1.super_16 1192913 106 - 1990440 AAUGGAAGCAGUUGUGUUGGAU---GAGUGUCCCGUUUAGGGUAAACAG-CAGCUAAAUGUUCGUAAA----UUCCCAUA-UUUAUGGACAUUUUAGCU-GUCAC--GUUUAUCAGUG (((((..(((.(..(.....).---.).))).)))))...(((((((.(-((((((((((((((((((----(......)-)))))))))).)))))))-))...--))))))).... ( -34.70, z-score = -4.16, R) >dp4.chr4_group4 838243 106 + 6586962 AAUGGAAGCAGUUGUGUUGGAU---GAGUGUCCCGUUUAGGGUAAACAG-CAGCUAAAUGUUCGUAAA----UUCCCAUA-UUUAUGGACAUUUUAGCU-GUCAC--GUUUAUCAGUG (((((..(((.(..(.....).---.).))).)))))...(((((((.(-((((((((((((((((((----(......)-)))))))))).)))))))-))...--))))))).... ( -34.70, z-score = -4.16, R) >droAna3.scaffold_12916 4346749 108 + 16180835 AAUGGAAGCCAUUGUGUC--GCUUGAAAUUCUAUGUUUAGGAUAAAUAC-CAGCUAAAUGUUUGUAGA----UUUUGCCA-UUUAUGGACAACUUAGCUGGCCAA--GUUUAUCAGUU (((((...))))).....--(((((..((((((....)))))).....(-(((((((.((((..((((----(......)-))))..))))..)))))))).)))--))......... ( -29.00, z-score = -2.32, R) >droEre2.scaffold_4929 15114266 110 + 26641161 AAUGGAAGCCAUUGUGCUGGGCCACGAACUCCAGGUUUAGGAUAAAUAG-CAGCUAACUGUUUGUAAA----UUUCACCA-UUUAUGGACAGUUUAGCUGGUCAC--GUUUAUCAGUG (((((...)))))...(((((((..........)))))))((((((((.-((((((((((((..((((----(......)-))))..))))).))))))).)...--))))))).... ( -34.20, z-score = -2.54, R) >droYak2.chr2L 14028978 110 - 22324452 AAUGGAAGCCAUUGUGCUGGGCCACGAACUCCAGGUUUAGGAUAAACAC-CAGCUAACUGUUUGUAAA----UUUCGCCA-UUUAUGGACAGCUUAGCUGGCCAC--GUUUAUCAGUG (((((...)))))...(((((((..........)))))))(((((((.(-((((((((((((..((((----(......)-))))..))))).))))))))....--))))))).... ( -38.20, z-score = -3.14, R) >droSec1.super_3 202066 110 - 7220098 AAUGGAAGCCAUUGUGCUGGGCCACGAACUCCAGGUUUAGGAUAAAUAC-CAGCUAACUGUUUGUAAA----UUUCGCCA-UUUAUGGACAGUUUAGCUGGCCAC--GUUUAUCAGUG (((((...)))))...(((((((..........)))))))(((((((.(-((((((((((((..((((----(......)-))))..))))).))))))))....--))))))).... ( -36.70, z-score = -2.98, R) >droSim1.chr2L 13699049 110 - 22036055 AAUGGAAGCCAUUGUGCUGGGCCACGAACUCCAGGUUUAGGAUAAAUAC-CAGCUAACUGUUUGUAAA----UUUCGCCA-UUUAUGGACAGUUUAGCUGGCCAC--GUUUAUCAGUG (((((...)))))...(((((((..........)))))))(((((((.(-((((((((((((..((((----(......)-))))..))))).))))))))....--))))))).... ( -36.70, z-score = -2.98, R) >consensus AAUGGAAGCCAUUGUGCUGGGC_ACGAACUCCAGGUUUAGGAUAAAUAC_CAGCUAACUGUUUGUAAA____UUUCGCCA_UUUAUGGACAGUUUAGCUGGCCAC__GUUUAUCAGUG ........................................(((((((...((((((((((((((((((.............))))))))))).))))))).......))))))).... (-19.21 = -18.39 + -0.82)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:39:04 2011