| Sequence ID | dm3.chr2L |
|---|---|
| Location | 13,929,511 – 13,929,615 |
| Length | 104 |
| Max. P | 0.585869 |

| Location | 13,929,511 – 13,929,615 |
|---|---|
| Length | 104 |
| Sequences | 12 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 81.23 |
| Shannon entropy | 0.39347 |
| G+C content | 0.42316 |
| Mean single sequence MFE | -24.94 |
| Consensus MFE | -15.67 |
| Energy contribution | -16.16 |
| Covariance contribution | 0.49 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.585869 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
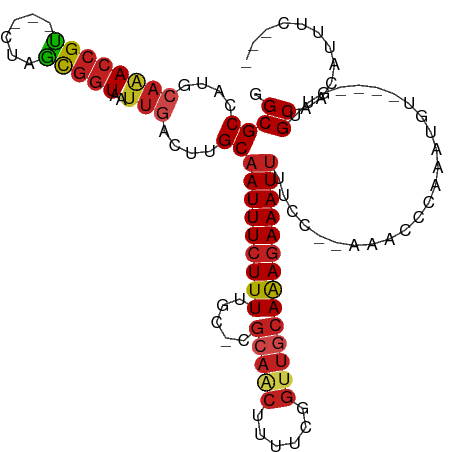
>dm3.chr2L 13929511 104 - 23011544 GGCGCCAUGCAAACCGU---CUAACGGUAAUUGACUUGCAAUUUCUUUUGCCCGCAACUUUUCGGUUGCAAAGAAAUUUUCC--AAACCCAAAUGU----GUUGCCACAUUUC--- ((((.(((((((((((.---....)))).......))))(((((((((.....((((((....)))))))))))))))....--..........))----).)))).......--- ( -25.41, z-score = -1.78, R) >droSim1.chr2L 13683785 104 - 22036055 GGCGCCAUGCAAACCGU---CUAGCGGUAAUUGACUUGCAAUUUCUUUUGCCCGCAACUUUUCGGUUGCAAAGAAAUUUUCC--AAACCCAAAUGU----GUUGCCACAUUUC--- ((((.(((((((((((.---....)))).......))))(((((((((.....((((((....)))))))))))))))....--..........))----).)))).......--- ( -25.71, z-score = -1.50, R) >droSec1.super_3 186763 104 - 7220098 GGCGCCAUGCAAACCGU---CUAGCGGUAAUUGACUUGCAAUUUCUUUUGCCCGCAACUUUUCGGUUGCAAAGAAAUUUUCC--AAACCCAAAUGU----GUUGCCACAUUUC--- ((((.(((((((((((.---....)))).......))))(((((((((.....((((((....)))))))))))))))....--..........))----).)))).......--- ( -25.71, z-score = -1.50, R) >droYak2.chr2L 14013376 103 - 22324452 GGCGCCAUGCAAACCGU---CUAGCGGUAAUUGACUUGCAAUUUCUUUUGCUCGCAACUUUUCGGUUGCAAAGAAAUUUUCC--AAACCCAAACA-----GUUGCCACAUUUC--- (((((....(((((((.---....))))..)))....))(((((((((.....((((((....)))))))))))))))....--...........-----...))).......--- ( -24.10, z-score = -1.44, R) >droEre2.scaffold_4929 15097954 104 + 26641161 GGCGCCAUGCAAACCGU---CUAGCGGUAAUUGACUUGCAAUUUCUUUUGCCCGCAACUUUUCGGUUGCAAAGAAAUUUUCC--AAACCCAAAUGU----GUUGCCACAUUUC--- ((((.(((((((((((.---....)))).......))))(((((((((.....((((((....)))))))))))))))....--..........))----).)))).......--- ( -25.71, z-score = -1.50, R) >droAna3.scaffold_12916 4331973 105 + 16180835 GGCGCCAUGCAAACCGU---CUAACGGUAAUUGACUUGCAAUUUCUUUUGCGCGCAGCUUUUCGGUUGCAAAGAAAUUUUCC-AAAACUCAAAUG----GGUUGCAACAUUUC--- .((((.......((((.---....)))).........((((......)))))))).........((((((((....)))...-..(((((....)----))))))))).....--- ( -25.90, z-score = -0.84, R) >dp4.chr4_group4 816230 107 + 6586962 GGCGCCAUGCAAACCGU---CUAAUGGUAAUUGACUUGCAAUUUCUUUCGC-CGCAACAUUUCGGCUGCAAAGAAAUUUUCC--AAACUCAAAUAUGUAAGUUGCAACAUUUC--- .(((((((.........---...)))))....(((((((((((((((((((-((........)))).).)))))))).....--...........))))))))))........--- ( -22.89, z-score = -1.09, R) >droPer1.super_16 1171202 107 - 1990440 GGCGCCAUGCAAACCGU---CUAAUGGUAAUUGACUUGCAAUUUCUUUCGC-CGCAACAUUUCGGCUGCAAAGAAAUUUUCC--AAACUCAAAUAUGUAAGUUGCAACAUUUC--- .(((((((.........---...)))))....(((((((((((((((((((-((........)))).).)))))))).....--...........))))))))))........--- ( -22.89, z-score = -1.09, R) >droWil1.scaffold_180708 10912292 95 + 12563649 GGCGCCGCCGCCACCGCC--UAAGCCGUAAUUGACUUGCAAUUUCUUUUGCAAACGAAAGAGAAAUAAAAGAAAAAU------------CAAAU------GUUGCAAGAGUUUGC- ((((....))))((.((.--...)).))......((((((((((((((((....))))))))).......((....)------------)....------.))))))).......- ( -21.80, z-score = -1.10, R) >droVir3.scaffold_12723 903261 96 - 5802038 GGCGCCAAGCAGACCGU---CUAGCGGUAAUUGACUUGCAAUUUCUUUUGC-AGCAACAUUUCGGUUGCAAAGAAAUUUAC--------CAAAUAU----GUUGCAAUAGUU---- .((((.(((((.((((.---....))))...)).)))))(((((((((.((-(((.........))))))))))))))...--------.......----...)).......---- ( -24.00, z-score = -1.06, R) >droMoj3.scaffold_6500 21913741 107 + 32352404 GGCGCCAACCAGACCGUCUACUAACGGUAAUUGACUUGCAAUUUCUUUUGC-GGCAACUUUUCGGUUGCAAAGAAAUUUUUCCAAAACACAAAUGU----GUUGCAAUAGCU---- (((..(((....(((((......)))))..)))..(((((((((((((.((-(((.........)))))))))))))).......(((((....))----)))))))..)))---- ( -27.20, z-score = -1.77, R) >droGri2.scaffold_15252 4410658 100 - 17193109 GGCGCCAAGCAGACCGU---CUAGCGUUAAUUUACCAGCAAUUUCUUUUGC-AGCAACUUUUUGGUUACAAAGAAAUUUGC--------CAAAUA----GGUUGCAAUAUGUGGUU (((((.(..(.....).---.).))))).....((((((((......))))-.((((((((((((...((((....)))))--------)))).)----))))))......)))). ( -28.00, z-score = -1.96, R) >consensus GGCGCCAUGCAAACCGU___CUAGCGGUAAUUGACUUGCAAUUUCUUUUGC_CGCAACUUUUCGGUUGCAAAGAAAUUUUCC__AAACCCAAAUGU____GUUGCAACAUUUC___ .((((....((((((((......)))))..)))....))(((((((((.....(((((......)))))))))))))).........................))........... (-15.67 = -16.16 + 0.49)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:39:01 2011