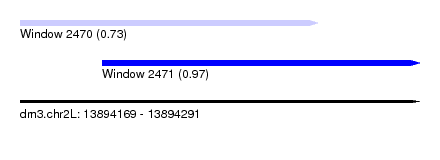
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 13,894,169 – 13,894,291 |
| Length | 122 |
| Max. P | 0.970498 |

| Location | 13,894,169 – 13,894,260 |
|---|---|
| Length | 91 |
| Sequences | 12 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 68.23 |
| Shannon entropy | 0.62906 |
| G+C content | 0.60194 |
| Mean single sequence MFE | -38.43 |
| Consensus MFE | -11.65 |
| Energy contribution | -11.75 |
| Covariance contribution | 0.10 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.30 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.729487 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2L 13894169 91 + 23011544 AAUGCACACAGCAACG-CAGACG--AG----UGUCCUCGUU---GCCAGGACAU--UCUAGCAG---CCAGUUGUUUGUCGACUGGCGUCGCGUGCGGGCGGCUUU----- ..(((.(...(((.((-(.((((--((----((((((....---...)))))))--)).....(---(((((((.....))))))))))))))))).)))).....----- ( -40.50, z-score = -2.76, R) >droSim1.chr2L 13646788 91 + 22036055 AAUGCACACAGCAAGG-CAGACG--AG----UGUCCUCGUU---GCCAGGACAU--UCUAGCAG---CCAGUUGUUUGUCGACUGGCGUCGCGUGCGGGCGGCUUU----- ..(((.(...(((..(-(.((((--((----((((((....---...)))))))--)).....(---(((((((.....))))))))))))).))).)))).....----- ( -38.10, z-score = -2.02, R) >droSec1.super_3 151135 91 + 7220098 AAUGCACACAGCAAGG-CAGACG--AG----UGUCCUCGUU---GCCAGGACAU--UCUAGCAG---CCAGUUGUUUGUCGACUGGCGUCGCGUGCGGGCGGCUUU----- ..(((.(...(((..(-(.((((--((----((((((....---...)))))))--)).....(---(((((((.....))))))))))))).))).)))).....----- ( -38.10, z-score = -2.02, R) >droYak2.chr2L 10300534 98 + 22324452 AAUGCACACAGCACACAGCAACGCAGACGAGUGUCCUCGUC---GCCAGGACAU--UCUAGCAG---CCAGUUGUUUGUCGACUGGCGUCGCGUGCGGGCGGCUUU----- ...((.....((.(...(((.(((.((((((((((((....---...)))))))--)).....(---(((((((.....))))))))))))))))).))).))...----- ( -41.30, z-score = -2.33, R) >droEre2.scaffold_4929 15062461 91 - 26641161 AAUGCACACAGCAACG-CAGACG--AG----UGUCCUCGUC---GCCAGGACAU--UCUAGCAG---CCAGUUGUUUGUCGACUGGCGUCGCGUGCGGGCGGCUUU----- ..(((.(...(((.((-(.((((--((----((((((....---...)))))))--)).....(---(((((((.....))))))))))))))))).)))).....----- ( -40.50, z-score = -2.71, R) >droAna3.scaffold_12916 4294847 98 - 16180835 AAUGCACACAGCAACACAGGACAGCAGACUGUGUCCUCGUU---GCCAGGAUGU--UCUGGCUG---CCAGUUGUUUGUCGACUGGCGUCGCGUGCGGGCGGCUUU----- ..(((.....)))..((((((((........)))))).)).---((((((....--)))))).(---(((((((.....))))))))(((((......)))))...----- ( -38.30, z-score = -0.65, R) >dp4.chr4_group4 776446 96 - 6586962 AAUGCACACAGCAACG-AGGACGCCGU----UGUCCUCGUCGUUGCCAGGACAU--UCUCGAAG---CCAGUUGUUUGUCGACUGGCGUCGCGUGCGGGCGGCUUU----- ..(((.....)))(((-((((((....----))))))))).((((((..(.(((--...(((.(---(((((((.....)))))))).))).)))).))))))...----- ( -41.80, z-score = -2.24, R) >droPer1.super_16 1099384 96 + 1990440 AAUGCACACAGCAACG-AGGACGCCGU----UGUCCUCGUCGUUGCCAGGACAU--UCUCGAAG---CCAGUUGUUUGUCGACUGGCGUCGCGUGCGGGCGGCUUU----- ..(((.....)))(((-((((((....----))))))))).((((((..(.(((--...(((.(---(((((((.....)))))))).))).)))).))))))...----- ( -41.80, z-score = -2.24, R) >droWil1.scaffold_180708 10863221 91 - 12563649 ---ACCGACUGUGGCAAAGGACGC--G-------CCGUGUU---GCCAGGAUAUAUUCUCGCUGUCGCCAGUUGUUUGUCGACUGGCUUGCCGUGCGGGCGGUUUU----- ---((((.(((((((((..(((((--(-------..(((((---.....))))).....))).)))((((((((.....)))))))))))))..)))).))))...----- ( -34.60, z-score = -0.82, R) >droMoj3.scaffold_6500 21870560 85 - 32352404 ----------------CAGGACGA--------GGCCGCAGGACAACGAGCGCGUCGACCAAGA--CGCCAGUUGUUUGUCGACUGGCAUUGCGGGCGGGCGGCUUUUCCAG ----------------..(((.((--------((((((.......((..((((((......))--)((((((((.....))))))))...)))..)).))))))))))).. ( -40.40, z-score = -3.19, R) >droGri2.scaffold_15252 4372928 81 + 17193109 ----------------CAGGACGA--------GGCCAACGGACAACGAGAGCGAUGAGCGAAAG-CGCCAGUUGUUUGUCGACUGGCGUCGCUGGCGGGCGGCUUU----- ----------------......((--------((((..((.....))..((((((..((....)-)((((((((.....)))))))))))))).......))))))----- ( -35.50, z-score = -2.35, R) >droVir3.scaffold_12723 856735 80 + 5802038 ----------------CAGGACGA--------GGCCAGAGGACGACGAGCGCGACGAGCCUGA--CGCCAGUUGUUUGUCGACUGGCAUCGCUGGCGGGCGGCAUU----- ----------------........--------.(((...(.....)..((.((.(.(((..((--.((((((((.....)))))))).))))).))).)))))...----- ( -30.20, z-score = -0.49, R) >consensus AAUGCACACAGCAACG_AGGACG__AG____UGUCCUCGUU___GCCAGGACAU__UCUAGCAG___CCAGUUGUUUGUCGACUGGCGUCGCGUGCGGGCGGCUUU_____ ...................................................................(((((((.....)))))))..((((......))))......... (-11.65 = -11.75 + 0.10)
| Location | 13,894,194 – 13,894,291 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 96.91 |
| Shannon entropy | 0.05018 |
| G+C content | 0.56742 |
| Mean single sequence MFE | -36.42 |
| Consensus MFE | -35.15 |
| Energy contribution | -35.40 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.96 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.83 |
| SVM RNA-class probability | 0.970498 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
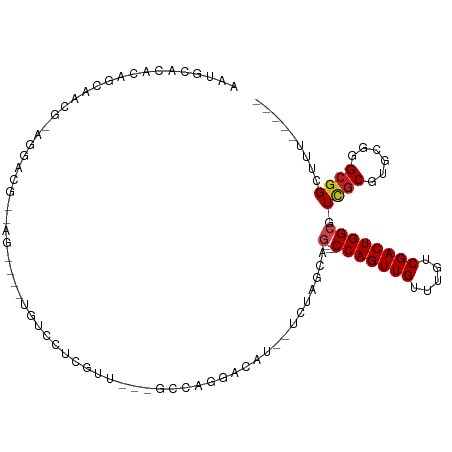
>dm3.chr2L 13894194 97 + 23011544 GUCCUCGUUGCCAGGACAUUCUAGCAGCCAGUUGUUUGUCGACUGGCGUCGCGUGCGGGCGGCUUUUUUUCCCAUUGCCGAAAGGCUGCUCCUUUUU (((((.......))))).....((((((((((((.....))))))))(((((......))))).............(((....)))))))....... ( -37.30, z-score = -2.46, R) >droSim1.chr2L 13646813 97 + 22036055 GUCCUCGUUGCCAGGACAUUCUAGCAGCCAGUUGUUUGUCGACUGGCGUCGCGUGCGGGCGGCUUUUUUUCCCAUCGCCGAAAGGCUGCUCCUUUUU (((((.......))))).....((((((((((((.....))))))))(((((......))))).............(((....)))))))....... ( -37.60, z-score = -2.48, R) >droSec1.super_3 151160 97 + 7220098 GUCCUCGUUGCCAGGACAUUCUAGCAGCCAGUUGUUUGUCGACUGGCGUCGCGUGCGGGCGGCUUUUUUUCCCAUCGCCGAAAGGCUGCUCCUUUUU (((((.......))))).....((((((((((((.....))))))))(((((......))))).............(((....)))))))....... ( -37.60, z-score = -2.48, R) >droEre2.scaffold_4929 15062486 95 - 26641161 GUCCUCGUCGCCAGGACAUUCUAGCAGCCAGUUGUUUGUCGACUGGCGUCGCGUGCGGGCGGCUUUUUU--CCAUCGCCGAACUGCUGCUCCUUUUU (((((.......))))).........((((((((.....))))))))...(((.((((.((((......--.....))))..)))))))........ ( -33.20, z-score = -1.42, R) >consensus GUCCUCGUUGCCAGGACAUUCUAGCAGCCAGUUGUUUGUCGACUGGCGUCGCGUGCGGGCGGCUUUUUUUCCCAUCGCCGAAAGGCUGCUCCUUUUU (((((.......))))).....((((((((((((.....))))))))(((((......))))).............(((....)))))))....... (-35.15 = -35.40 + 0.25)

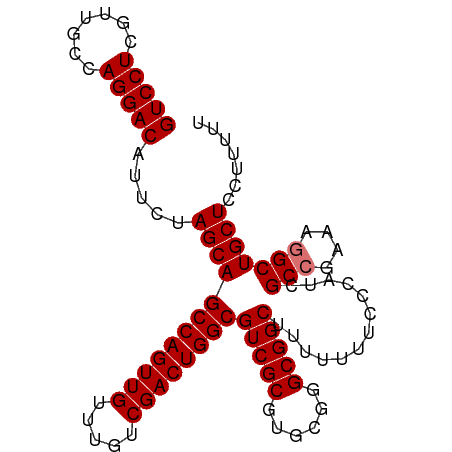
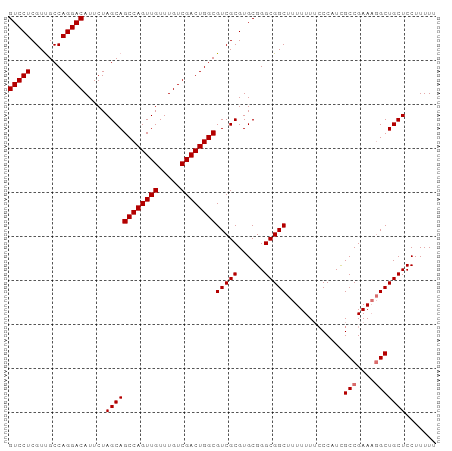
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:38:59 2011