
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 13,813,146 – 13,813,250 |
| Length | 104 |
| Max. P | 0.812512 |

| Location | 13,813,146 – 13,813,250 |
|---|---|
| Length | 104 |
| Sequences | 7 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 61.14 |
| Shannon entropy | 0.78692 |
| G+C content | 0.45674 |
| Mean single sequence MFE | -30.20 |
| Consensus MFE | -10.66 |
| Energy contribution | -10.39 |
| Covariance contribution | -0.27 |
| Combinations/Pair | 1.95 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.812512 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
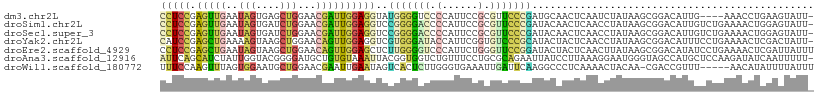
>dm3.chr2L 13813146 104 + 23011544 CCUCCGAGUUGAAUAGUGAGCUGGAACGAUUGGAGGUAUGGGGUCCCCAUUCCGCGUUCCCGAUGCAACUCAAUCUAUAAGCGGACAUUG----AAACCUGAAGUAUU- (((((.(((((..(((....)))...))))))))))...((..((.....(((((......(((........))).....)))))....)----)..)).........- ( -25.70, z-score = 0.27, R) >droSim1.chr2L 13572948 108 + 22036055 CCUCCGAGUUGAAUAGUGAUCUGGAACGAUUGGAGGUCCGGGGACCCCAUUCCGCGUUCCCGAUACAACUCAACCUAUAAGCGGACAUUGUCUGAAAACUGGAGUAUU- .((((((((((.......(((.((((((..(((.((((....))))))).....)))))).))).))))))..........(((((...)))))......))))....- ( -38.10, z-score = -2.69, R) >droSec1.super_3 78644 108 + 7220098 CCUCCGAGUUGAAUAGUGAUCUGGAACGAUUGGAGGUCCGGGGACCCCAUUCCGCGUUCCCGAUACAACUCAACCUAUAAGCGGACAUUGUCUGAAAACUGGAGUAUU- .((((((((((.......(((.((((((..(((.((((....))))))).....)))))).))).))))))..........(((((...)))))......))))....- ( -38.10, z-score = -2.69, R) >droYak2.chr2L 10225025 108 + 22324452 CAUCCGAGCUGAAAAGUAAGCUGGAACAGUUGGAGGUCGUGGGAUACCAUUCGGUGUCCCGCAUACUACUCAACCUAUAAGCGGACAUUUCCUGAAAACUCGACUAUU- ..(((.((((........)))))))..(((((.((.((((((((((((....))))))))))....................(((....))).))...)))))))...- ( -35.00, z-score = -2.88, R) >droEre2.scaffold_4929 14988299 109 - 26641161 CCUCCGAGCUGAAUAGUAAGCUGGAACAGUUGGAGCUCUUGGGGUCCCAUUCUGGGUUCCGGAUACUACUCAACUUAUAAGCGGACAUAUCCUGAAAACUCGAUUAUUU ....((((.(((.(((((..(((((((...(((..((.....))..)))......))))))).))))).)))..........(((....)))......))))....... ( -29.10, z-score = -0.52, R) >droAna3.scaffold_12916 4214542 108 - 16180835 AUUCAGCAUCUAUUGGUACGGGGAUGCUGUGUAAAUUACGGUGGUCUGUUUCCUGCGCAGAAUUAUCCUUAAAGGAAUGGGUAGCCAUGCUCCAAGAUAUCAAUUUUU- ...........((((((((((((((((((((((....((((....))))....)))))))...)))))))...(((((((....))))..)))..).)))))))....- ( -27.30, z-score = -0.44, R) >droWil1.scaffold_180772 7128418 103 + 8906247 UUUCCAAGUUUAGUGGAAUGCUGGAACGAAUUGAAUAGUCACUCUUGGGUGAAAUUGAUUCAAGGCCCUCAAAACUACAA-CGACCGUUU-----AACAUAUUUUAUUU .......(((((((.((..(((.....((((..(....(((((....)))))..)..))))..)))..))...)))).))-)........-----.............. ( -18.10, z-score = 0.27, R) >consensus CCUCCGAGUUGAAUAGUAAGCUGGAACGAUUGGAGGUCCGGGGAUCCCAUUCCGCGUUCCCGAUACAACUCAACCUAUAAGCGGACAUUGUCUGAAAACUCGAGUAUU_ (((((.(((((..(((....)))...))))))))))..(((((((.(......).)))))))............................................... (-10.66 = -10.39 + -0.27)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:38:53 2011