
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 13,772,896 – 13,772,992 |
| Length | 96 |
| Max. P | 0.691129 |

| Location | 13,772,896 – 13,772,992 |
|---|---|
| Length | 96 |
| Sequences | 12 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 63.37 |
| Shannon entropy | 0.81519 |
| G+C content | 0.42464 |
| Mean single sequence MFE | -19.22 |
| Consensus MFE | -8.71 |
| Energy contribution | -8.82 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.56 |
| Mean z-score | -0.44 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.691129 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
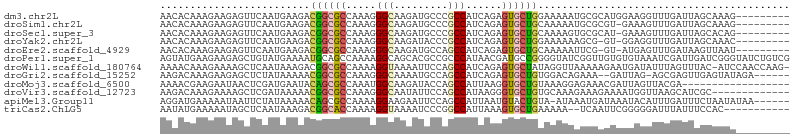
>dm3.chr2L 13772896 96 - 23011544 AACACAAAGAAGAGUUCAAUGAAGACGGCGCCAAAGGGCAAGAUGCCCGCCAUCAGAGUGCUGGAAAAAUGCGCAUGGAAGGUUUGAUUAGCAAAG--------- ...........................((......((((.....))))(((.((...((((.........))))...)).))).......))....--------- ( -19.50, z-score = 0.66, R) >droSim1.chr2L 13532114 95 - 22036055 AACACAAAGAAGAGUUCAAUGAAGACGGCGCCAAAGGGCAAGAUGCCCGCCAUCAGAGUGCUGCAAAAAUGCGCGU-GAAAGUUUGAUUAGCAAAG--------- .............(((.(((..(((((((......((((.....))))))).(((..((((.........)))).)-))..)))).))))))....--------- ( -20.50, z-score = 0.24, R) >droSec1.super_3 38729 95 - 7220098 AACACAAAGAAGAGUUCAAUGAAGACGGCGCCAAAGGGCAAGAUGCCCGCCAUCAGAGUGCUGCAAAAGUGCGCAU-GAAAGUUUGAUUAGCACAG--------- .............(((.(((..(((((((......((((.....))))))).(((..(..((.....))..)...)-))..)))).))))))....--------- ( -21.20, z-score = 0.08, R) >droYak2.chr2L 10184650 94 - 22324452 AACACAAAGAAGAGUUCAAUGAAGACGGCGCCAAAGGGCAAGAUACCCGCCAUCAGAGUGCUGGAAAAAAGCG-GU-GGAGGUUUGAUUAGCAAAC--------- ..(((..................((.((((((....)))..(.....)))).))....((((.......))))-))-)...(((((.....)))))--------- ( -17.50, z-score = 0.60, R) >droEre2.scaffold_4929 14947822 94 + 26641161 AACACAAAGAAGAGUUCAAUGAAGACGGCGCCAAAGGGCAAGAUGCCAGCCAUCAGAGUGCUGCAAAAAUUCG-GU-AUGAGUUUGAUAAGUUAAU--------- .......................((((((.......(((.....))).)))((((((((((((........))-))-))...))))))..)))...--------- ( -16.90, z-score = 0.38, R) >droPer1.super_1 9008798 105 - 10282868 AGUAUGAAGAAGAGCUGUAUGAAAAUGCAGCCAAAAGGCAGCACGCCGCCCAUAACGAUGCCGGGGUAUCGGUUGUGUGUAAAUCGAUUGAUCGGGUAUCUGUCG ..((((.....(.(((((((....))))))))....(((.....)))...))))..((((((.((..(((((((.......)))))))...)).))))))..... ( -35.20, z-score = -2.25, R) >droWil1.scaffold_180764 3904831 103 + 3949147 AAAACAAAGAAAAGCUCAAUAAAGACGGCGCCAAAAGGUAAAAUUCCAGCCAUCAGAGUGCUAUAGGUUAAAAAGAAUGAUAUUUAGUUUAC-AUCCAACCAAG- .......((....((((......((.((((((....))).........))).)).)))).))...((((.....((((........))))..-....))))...- ( -10.10, z-score = 0.85, R) >droGri2.scaffold_15252 4244973 96 - 17193109 AAGACAAAGAAGAGCUCUAUAAAAACGGCGCCAAAGGGCAAAAUGCCAGCCAUCAGAGUGCUGUGGACAGAAA--GAUUAG-AGCGAGUUGAGUAUAGA------ ....(((......((((((.......((((((....))).....))).((((.(((....)))))).).....--...)))-)))...)))........------ ( -21.60, z-score = -1.31, R) >droMoj3.scaffold_6500 29221415 87 - 32352404 AAAACGAAGAAUAACUCGAUGAAUACAGCGCCAAAUGGCAAGAUACCAGCCAUUAAGGUGCUGUAAAGGAGAAACGAUUAGUUACGA------------------ ....((........(((..(...(((((((((.((((((.........))))))..))))))))).).))).(((.....))).)).------------------ ( -23.70, z-score = -4.39, R) >droVir3.scaffold_12723 703415 92 - 5802038 AAGACAAAGAAAAGCUCGAUAAAAACGGCGCCAAAGGGCAAUAUUCCAGCCAUAAGGGUGCUGUGCAAAGAAAGAAAAUGGUUAAGCAUCGC------------- .............(((..((....((((((((....(((.........))).....))))))))................))..))).....------------- ( -21.25, z-score = -1.25, R) >apiMel3.Group11 8200905 98 + 12576330 AGGAUGAAAAAUAAUUCUAUAAAAACAGCGCCAAAAGGAAGAAUUCCAGCCAUUAAUGUACUGUA-AUAAAUGAUAAAUACAUUUGAUUUCUAAUAUAA------ (((((........)))))......((((..(.....((..(.....)..))......)..)))).-.((((((.......)))))).............------ ( -8.30, z-score = 0.90, R) >triCas2.ChLG5 3305218 92 + 18847211 AAUAUGAAAAAUAGCUCAAUAAAGACGGCACCAAAAGGUAAAAUCCCGGCCAUUAAAGUGCUGAAAAA--UCAAUUCGGGGGAUUUAUUUCCAC----------- .....((((....(((..........)))(((....))).(((((((...(((....)))(((((...--....)))))))))))).))))...----------- ( -14.90, z-score = 0.18, R) >consensus AACACAAAGAAGAGCUCAAUGAAGACGGCGCCAAAGGGCAAGAUGCCAGCCAUCAGAGUGCUGUAAAAAAGCGAGUAGUAGGUUUGAUUAGCAAAG_________ .........................((((((.....(((.........)))......)))))).......................................... ( -8.71 = -8.82 + 0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:38:49 2011