| Sequence ID | dm3.chr2L |
|---|---|
| Location | 13,747,739 – 13,747,830 |
| Length | 91 |
| Max. P | 0.999801 |

| Location | 13,747,739 – 13,747,830 |
|---|---|
| Length | 91 |
| Sequences | 9 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 78.58 |
| Shannon entropy | 0.43367 |
| G+C content | 0.35477 |
| Mean single sequence MFE | -21.86 |
| Consensus MFE | -16.49 |
| Energy contribution | -16.80 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.73 |
| SVM RNA-class probability | 0.994698 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
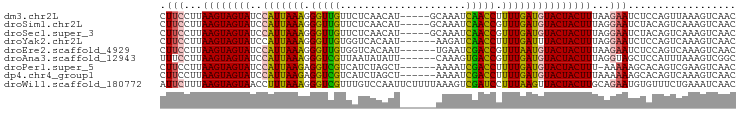
>dm3.chr2L 13747739 91 + 23011544 GUUGACUUUAACUGGAGAUUCUUAAAGUAGUACAUCAAAAGGUUGAUUUGC-----AUGUUGAGAACAACCCUUUAAUGGAUACUACUUAAGGAAG .....((((....)))).(((((.(((((((((((.(((.(((((.(((..-----......))).))))).))).)))..)))))))).))))). ( -20.70, z-score = -1.79, R) >droSim1.chr2L 13506676 91 + 22036055 GUUGACUUUGACUGUAGAUUCCUAAAGUAGUACAUCAAACGGUUGAUUUGC-----AUGUUGAGAACAACCCUUUAAUGGAUACUACUUAAGGAAG ..................(((((.(((((((((((.(((.(((((.(((..-----......))).))))).))).)))..)))))))).))))). ( -23.00, z-score = -2.56, R) >droSec1.super_3 14603 91 + 7220098 GUUGACUUUGACUGUAGAUUCCUAAAGUAGUACAUCAAACGGUUGAUUUGC-----AUGUUGAGAACAACCCUUUAAUGGAUACUACUUAAGGAAG ..................(((((.(((((((((((.(((.(((((.(((..-----......))).))))).))).)))..)))))))).))))). ( -23.00, z-score = -2.56, R) >droYak2.chr2L 10160058 90 + 22324452 GUUGACUUUGACUGGAGAUUCCUAAAGUAGUAAAUCAAAAGGUUGAUCUU------AUUGUGACCACAACCCUUUAAUGGAUACUACUUAAGGAAG .....((((....)))).(((((.((((((((.((.(((.(((((.((..------.....))...))))).))).))...)))))))).))))). ( -21.00, z-score = -1.70, R) >droEre2.scaffold_4929 14922150 90 - 26641161 GUUGACUUUGACUGGAGAUUCUUAAAGUAGUACAUUAAACGGUCGAUUCA------AUUGUGACCACAACCCUUUAAUGGAUACUACUUAAGGAAG .....((((....)))).(((((.(((((((((((((((.(((.(..(((------....)))...).))).)))))))..)))))))).))))). ( -21.00, z-score = -1.87, R) >droAna3.scaffold_12943 1815546 90 + 5039921 GCCGACUUUAAAUGGAGCUACCUAAAGUAGUACAUCAAACGGUCACUUUG------AAUAUAUUAACGACCCUUUAAUGGAUACUACUUAAGGAAA .(((........))).....(((.(((((((((((.(((.((((...(((------(.....)))).)))).))).)))..)))))))).)))... ( -18.90, z-score = -2.32, R) >droPer1.super_5 3483604 89 + 6813705 GUUGACUUCGACUGUGCUUUUU-AAAGUAGUACAUCAAAAGGUCGAUUUU------AGCUAGAUGACGACCUCUUAAUGGAUACUACUUAAGGAAG ((((....))))....((((((-.(((((((((((.((.((((((((((.------....))))..)))))).)).)))..)))))))).)))))) ( -23.30, z-score = -2.41, R) >dp4.chr4_group1 3555999 90 - 5278887 GUUGACUUUGACUGUGCUUUUUUAAAGUAGUACAUCAAAAGGUCGAUUUU------AGCUAGAUGACGACCUCUUAAUGGAUACUACUUAAGGAAG ................(((((((((.(((((((((.((.((((((((((.------....))))..)))))).)).)))..))))))))))))))) ( -20.80, z-score = -1.71, R) >droWil1.scaffold_180772 8360837 96 - 8906247 GUUGAUUUCAGAAACACAUUCUGCAAGUAGUAACUUAAAGGAUCGACUUUAAAAGAAUUGGACAAACGACCCUUUAAAGGUUACUACUUAAAGAAU .................(((((..(((((((((((((((((.(((..(((((.....)))))....))).))))))..)))))))))))..))))) ( -25.00, z-score = -4.09, R) >consensus GUUGACUUUGACUGGAGAUUCUUAAAGUAGUACAUCAAAAGGUCGAUUUG______AUUUUGAGAACAACCCUUUAAUGGAUACUACUUAAGGAAG ..................(((((.(((((((((((.(((.(((((.....................))))).))).)))..)))))))).))))). (-16.49 = -16.80 + 0.31)


| Location | 13,747,739 – 13,747,830 |
|---|---|
| Length | 91 |
| Sequences | 9 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 78.58 |
| Shannon entropy | 0.43367 |
| G+C content | 0.35477 |
| Mean single sequence MFE | -25.97 |
| Consensus MFE | -19.25 |
| Energy contribution | -18.58 |
| Covariance contribution | -0.67 |
| Combinations/Pair | 1.30 |
| Mean z-score | -4.76 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.43 |
| SVM RNA-class probability | 0.999801 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 13747739 91 - 23011544 CUUCCUUAAGUAGUAUCCAUUAAAGGGUUGUUCUCAACAU-----GCAAAUCAACCUUUUGAUGUACUACUUUAAGAAUCUCCAGUUAAAGUCAAC ....(((((((((((..(((((((((((((..........-----......))))))))))))))))))).))))).................... ( -23.69, z-score = -4.57, R) >droSim1.chr2L 13506676 91 - 22036055 CUUCCUUAAGUAGUAUCCAUUAAAGGGUUGUUCUCAACAU-----GCAAAUCAACCGUUUGAUGUACUACUUUAGGAAUCUACAGUCAAAGUCAAC .(((((.((((((((..(((((((.(((((..........-----......))))).))))))))))))))).))))).................. ( -27.19, z-score = -5.24, R) >droSec1.super_3 14603 91 - 7220098 CUUCCUUAAGUAGUAUCCAUUAAAGGGUUGUUCUCAACAU-----GCAAAUCAACCGUUUGAUGUACUACUUUAGGAAUCUACAGUCAAAGUCAAC .(((((.((((((((..(((((((.(((((..........-----......))))).))))))))))))))).))))).................. ( -27.19, z-score = -5.24, R) >droYak2.chr2L 10160058 90 - 22324452 CUUCCUUAAGUAGUAUCCAUUAAAGGGUUGUGGUCACAAU------AAGAUCAACCUUUUGAUUUACUACUUUAGGAAUCUCCAGUCAAAGUCAAC .(((((.((((((((...((((((((((..(((((.....------..))))))))))))))).)))))))).))))).................. ( -28.40, z-score = -4.92, R) >droEre2.scaffold_4929 14922150 90 + 26641161 CUUCCUUAAGUAGUAUCCAUUAAAGGGUUGUGGUCACAAU------UGAAUCGACCGUUUAAUGUACUACUUUAAGAAUCUCCAGUCAAAGUCAAC ....(((((((((((..(((((((.(((((...(((....------)))..))))).))))))))))))).))))).................... ( -25.00, z-score = -4.10, R) >droAna3.scaffold_12943 1815546 90 - 5039921 UUUCCUUAAGUAGUAUCCAUUAAAGGGUCGUUAAUAUAUU------CAAAGUGACCGUUUGAUGUACUACUUUAGGUAGCUCCAUUUAAAGUCGGC ...(((.((((((((..(((((((.(((((..........------.....))))).))))))))))))))).))).................... ( -24.16, z-score = -3.48, R) >droPer1.super_5 3483604 89 - 6813705 CUUCCUUAAGUAGUAUCCAUUAAGAGGUCGUCAUCUAGCU------AAAAUCGACCUUUUGAUGUACUACUUU-AAAAAGCACAGUCGAAGUCAAC (((....((((((((..(((((((((((((..........------.....))))))))))))))))))))).-...)))................ ( -24.46, z-score = -4.39, R) >dp4.chr4_group1 3555999 90 + 5278887 CUUCCUUAAGUAGUAUCCAUUAAGAGGUCGUCAUCUAGCU------AAAAUCGACCUUUUGAUGUACUACUUUAAAAAAGCACAGUCAAAGUCAAC .......((((((((..(((((((((((((..........------.....)))))))))))))))))))))........................ ( -23.86, z-score = -4.91, R) >droWil1.scaffold_180772 8360837 96 + 8906247 AUUCUUUAAGUAGUAACCUUUAAAGGGUCGUUUGUCCAAUUCUUUUAAAGUCGAUCCUUUAAGUUACUACUUGCAGAAUGUGUUUCUGAAAUCAAC (((((.(((((((((((..((((((((((((((..............))).)))))))))))))))))))))).)))))................. ( -29.74, z-score = -5.96, R) >consensus CUUCCUUAAGUAGUAUCCAUUAAAGGGUUGUUAUCAAAAU______AAAAUCGACCGUUUGAUGUACUACUUUAAGAAUCUACAGUCAAAGUCAAC .(((...((((((((..(((((((.(((((.....................))))).)))))))))))))))...))).................. (-19.25 = -18.58 + -0.67)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:38:46 2011