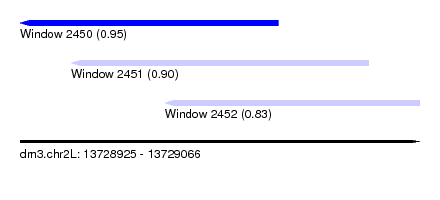
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 13,728,925 – 13,729,066 |
| Length | 141 |
| Max. P | 0.953148 |

| Location | 13,728,925 – 13,729,016 |
|---|---|
| Length | 91 |
| Sequences | 12 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 69.30 |
| Shannon entropy | 0.64856 |
| G+C content | 0.41100 |
| Mean single sequence MFE | -19.50 |
| Consensus MFE | -11.90 |
| Energy contribution | -11.82 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.07 |
| Mean z-score | -0.93 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.953148 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 13728925 91 - 23011544 AUAAUCUUAUUGACGUGAUAGGAUUACACUUUGCUAAGUGAAUUAAUAAAGUA-UCCUGUCAAGGGGGUUGGGGCGGAGGGGGGAUGUGGCC--------------- .(((((((.......(((((((((...((((((.(((.....))).)))))))-))))))))..)))))))((.((.(.......).)).))--------------- ( -21.00, z-score = -0.67, R) >droSim1.chr2L 13487510 83 - 22036055 AUAAUCUUAUUGACGUGAUAGGAUUACACUUUGCUAAGUGAAUUAAUAAAGUA-UCCUGAAAGGGGG--------GGGGGGGGGAUGUGACC--------------- .((((((((((.....))))))))))(((((....))))).............-((((....)))).--------.....((........))--------------- ( -16.40, z-score = -0.32, R) >droSec1.super_16 1856341 78 - 1878335 AUAAUCUUAUUGACGUGAUAGGAUUACACUUUGCUAAGUGAAUUAAUAAAGUA-UCCGGAC-------------CGAGGGGGGGAUGUGACC--------------- .((((((((((.....))))))))))(((((....)))))...........((-(((...(-------------(....)).))))).....--------------- ( -17.40, z-score = -0.80, R) >droYak2.chr2L 10140107 106 - 22324452 AUAAUCUUAUUGACGUGAUAGGAUUACACUUUGCUAAGUGAAUUAAUAAAGUA-ACCGGGCGGUGGGACAAGUUGGGACGGGGAAUUAUGAGGGAGGAAAUGUGGCC .((((((((((.....))))))))))((((((.((.....((((..(...((.-.((.(((..........))).)))).)..)))).......)).))).)))... ( -17.00, z-score = 0.90, R) >droAna3.scaffold_12943 1788744 97 - 5039921 AUAAUCUUAUUGACGUGAUAGGAUUACACUUUGCUAAGUGAAUUAAUAAAGUAUGCGAGCUGGAUAG-------GGAGCGACGAGUUUGGCCGAGAGUGUGGCC--- .((((((((((.....))))))))))(((((....)))))................(((((.(...(-------....)..).)))))(((((......)))))--- ( -22.00, z-score = -0.79, R) >droEre2.scaffold_4929 14903346 83 + 26641161 AUAAUCUUAUUGACGUGAUAGGAUUACACUUUGCUAAGUGAAUUAAUAAAGUA-ACCGGGCGGCGGG--------ACGGGGGAAAUGUGGCC--------------- .((((((((((.....))))))))))(((((....))))).............-....(((.(((..--------.(....)...))).)))--------------- ( -17.60, z-score = -0.56, R) >dp4.chr4_group1 3534556 82 + 5278887 AUAAUCUUAUUGACGUGAUAGGAUUACACUUUGCUAAGUGAAUUAAUAAAGUAUGCGAGUUUGCU----------GGCCAGAAAGUGUGGCC--------------- .((((((((((.....))))))))))(((((....))))).........((((........))))----------(((((.......)))))--------------- ( -19.60, z-score = -1.67, R) >droPer1.super_5 3462160 82 - 6813705 AUAAUCUUAUUGACGUGAUAGGAUUACACUUUGCUAAGUGAAUUAAUAAAGUAUGCGAGUUUGCU----------GGCCAGAAAGUGUGGCC--------------- .((((((((((.....))))))))))(((((....))))).........((((........))))----------(((((.......)))))--------------- ( -19.60, z-score = -1.67, R) >droWil1.scaffold_180772 8326832 79 + 8906247 AUAAUCUUAUUGACGUGAUAGGAUUACACUUUGCUAAGUGAAUUAAUAAAGUAUGCAGGAUGGUGGCAAAAGUGUGGCC---------------------------- .((((((((((.....))))))))))(((((....))))).....................(((.(((....))).)))---------------------------- ( -17.10, z-score = -1.26, R) >droVir3.scaffold_12723 724713 92 + 5802038 AUAAUCUUAUUGACGUGAUAGGAUUACACUUUGCUAAGUGAAUUAAUAAAGUAUGCGUCGCCGAUGUUGAUGCUGAUGCUGAGUUUGUGGCC--------------- .((((((((((.....))))))))))......((((...(((((.....(((((((((((.......))))))..))))).))))).)))).--------------- ( -20.50, z-score = -0.97, R) >droMoj3.scaffold_6500 16629923 90 - 32352404 AUAAUCCUAUUGACGUGAUAGGAUUACACUUUGCUAAGUGAAUUAAUAAAGUGUGUGAGCAUCGCAUCGCA--UAGCAUCGACUUGGCGGCC--------------- .((((((((((.....))))))))))....(((((((((((.((....)).(((((((((...)).)))))--))...)).)))))))))..--------------- ( -24.40, z-score = -1.41, R) >droGri2.scaffold_15126 307201 80 + 8399593 AUAAUCUUAUUGACGUGAUAGGAUUACACUUUGCUAAGUGAAUUAAUAAAGUAUGCGUUGUUGACGCCGCAUCGGUGGCC--------------------------- .((((((((((.....))))))))))(((((....)))))..(((((((.(....).))))))).(((((....))))).--------------------------- ( -21.40, z-score = -1.93, R) >consensus AUAAUCUUAUUGACGUGAUAGGAUUACACUUUGCUAAGUGAAUUAAUAAAGUAUGCGAGGCGGAGGG________GGGCGGGGAAUGUGGCC_______________ .((((((((((.....))))))))))(((((....)))))................................................................... (-11.90 = -11.82 + -0.08)
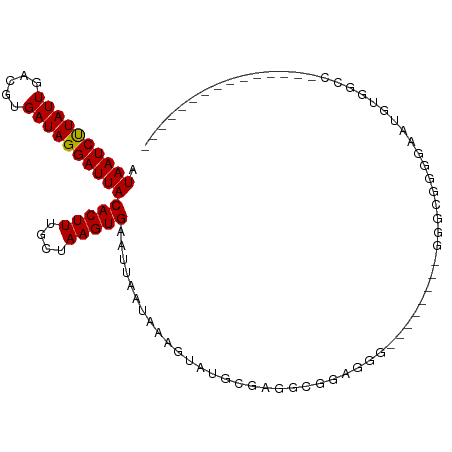
| Location | 13,728,943 – 13,729,048 |
|---|---|
| Length | 105 |
| Sequences | 12 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 80.40 |
| Shannon entropy | 0.40699 |
| G+C content | 0.32392 |
| Mean single sequence MFE | -18.03 |
| Consensus MFE | -14.00 |
| Energy contribution | -13.93 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.895437 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 13728943 105 - 23011544 CGGGAGGAAAAAUAUUAAAGAUUCUG------GAAACCAUAAUCUUAUUGACGUGAUAGGAUUACACUUUGCUAAGUGAAUUAAUAAAGUAUCCUGUCAAGGGGGUUGGGG------ (((((................)))))------....((.(((((((.......(((((((((...((((((.(((.....))).)))))))))))))))..))))))))).------ ( -23.09, z-score = -1.29, R) >droSim1.chr2L 13487528 87 - 22036055 ----------AAUAUUAAAGAUUCUG------GAAACCAUAAUCUUAUUGACGUGAUAGGAUUACACUUUGCUAAGUGAAUUAAUAAAGUAUCCUGAAAGGGG-------------- ----------..((((((.......(------....)..((((((((((.....))))))))))(((((....)))))..))))))......(((....))).-------------- ( -17.90, z-score = -1.79, R) >droSec1.super_16 1856359 82 - 1878335 ----------AAUAUUAAAGAUUCUG------GAAACCAUAAUCUUAUUGACGUGAUAGGAUUACACUUUGCUAAGUGAAUUAAUAAAGUAUCCGGAC------------------- ----------............((((------((.((..((((((((((.....))))))))))(((((....)))))..........)).)))))).------------------- ( -20.30, z-score = -3.55, R) >droYak2.chr2L 10140136 109 - 22324452 CAGAAGGAAAAAUAUUAAAGAUUCUG------GAAACCAUAAUCUUAUUGACGUGAUAGGAUUACACUUUGCUAAGUGAAUUAAUAAAGUAACCGGGCGGUGGGACAAGUUGGGA-- (((((................)))))------....(((((((((((((.....))))))))))(((((....)))))..........((.(((....)))...))....)))..-- ( -21.09, z-score = -0.92, R) >droAna3.scaffold_12943 1788762 111 - 5039921 CAACAAGAAAAAUAUUAAAGAUUCCG------AAAACCAUAAUCUUAUUGACGUGAUAGGAUUACACUUUGCUAAGUGAAUUAAUAAAGUAUGCGAGCUGGAUAGGGAGCGACGAGU ...................(.((((.------....(((((((((((((.....))))))))))(((((....)))))....................)))...)))).)....... ( -17.30, z-score = -0.32, R) >droEre2.scaffold_4929 14903361 100 + 26641161 CAGCAGGAAAAAUAUUAAAGAUUCUG------GAAACCAUAAUCUUAUUGACGUGAUAGGAUUACACUUUGCUAAGUGAAUUAAUAAAGUAACCGGGCGGCGGGAC----------- ..((.((.....((((((.......(------....)..((((((((((.....))))))))))(((((....)))))..))))))......))..))........----------- ( -17.70, z-score = -0.66, R) >dp4.chr4_group1 3534574 96 + 5278887 CAAGAGGAAAAAUAUUAAAGAUUCUG------AAAACCAUAAUCUUAUUGACGUGAUAGGAUUACACUUUGCUAAGUGAAUUAAUAAAGUAUGCGAGUUUGC--------------- ..................((((((..------.......((((((((((.....))))))))))(((((....)))))................))))))..--------------- ( -15.30, z-score = -0.88, R) >droPer1.super_5 3462178 96 - 6813705 CAAGAGGAAAAAUAUUAAAGAUUCUG------AAAACCAUAAUCUUAUUGACGUGAUAGGAUUACACUUUGCUAAGUGAAUUAAUAAAGUAUGCGAGUUUGC--------------- ..................((((((..------.......((((((((((.....))))))))))(((((....)))))................))))))..--------------- ( -15.30, z-score = -0.88, R) >droWil1.scaffold_180772 8326847 98 + 8906247 UAACAAGAAAAAUAUUAAAGAUUCUC----GAAAAACCAUAAUCUUAUUGACGUGAUAGGAUUACACUUUGCUAAGUGAAUUAAUAAAGUAUGCAGGAUGGU--------------- ..........................----.....((((((((((((((.....))))))))).(((((....)))))...................)))))--------------- ( -14.90, z-score = -0.92, R) >droVir3.scaffold_12723 724743 94 + 5802038 CAACAAGAAAAAUAUUAAAGAUUCUC------AAAACCAUAAUCUUAUUGACGUGAUAGGAUUACACUUUGCUAAGUGAAUUAAUAAAGUAUGCG-UCGCC---------------- ......((...(((((..........------.......((((((((((.....))))))))))(((((....))))).........)))))...-))...---------------- ( -15.00, z-score = -1.30, R) >droMoj3.scaffold_6500 16629942 108 - 32352404 -AACAAGAAAAAUAUUAAAGAUUCUCUCUCUUAAAACCAUAAUCCUAUUGACGUGAUAGGAUUACACUUUGCUAAGUGAAUUAAUAAAGUGUGUGAGCAUCGCAUCGCA-------- -...........((((((.....................((((((((((.....))))))))))(((((....)))))..))))))..(((((((.....)))).))).-------- ( -23.10, z-score = -2.34, R) >droGri2.scaffold_15126 307220 93 + 8399593 CAACAAGAAAAAUAUUAAAGAUUCUC------AAAACCAUAAUCUUAUUGACGUGAUAGGAUUACACUUUGCUAAGUGAAUUAAUAAAGUAUGCG-UUGU----------------- ((((........((((((........------.......((((((((((.....))))))))))(((((....)))))..))))))..(....))-))).----------------- ( -15.40, z-score = -1.41, R) >consensus CAACAAGAAAAAUAUUAAAGAUUCUG______AAAACCAUAAUCUUAUUGACGUGAUAGGAUUACACUUUGCUAAGUGAAUUAAUAAAGUAUGCGGGCGGGG_______________ ............((((((.....................((((((((((.....))))))))))(((((....)))))..))))))............................... (-14.00 = -13.93 + -0.08)

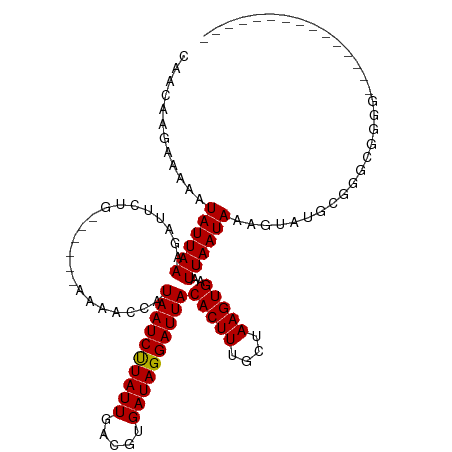
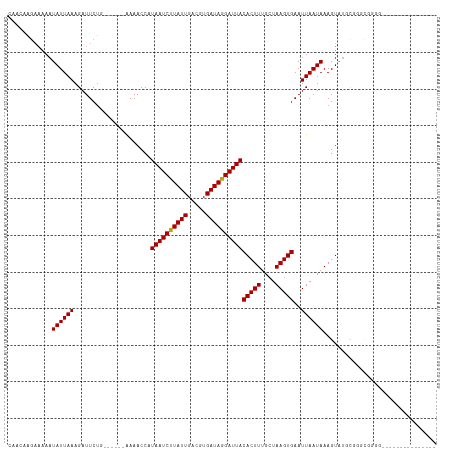
| Location | 13,728,976 – 13,729,066 |
|---|---|
| Length | 90 |
| Sequences | 11 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 75.85 |
| Shannon entropy | 0.51186 |
| G+C content | 0.36323 |
| Mean single sequence MFE | -17.53 |
| Consensus MFE | -10.83 |
| Energy contribution | -10.75 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.825907 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 13728976 90 - 23011544 ---------GCAUGUGUCUGCUACAAGCGGGAGGAAAAAUAUUAAAGAUUCU------GGAAACCAUAAUCUUAUUGACGUGAUAGGAUUACACUUUGCUAAGUG ---------.(((((.(((((.....))))).............((((((.(------((...))).))))))....))))).........(((((....))))) ( -18.30, z-score = -0.92, R) >droSim1.chr2L 13487553 79 - 22036055 ---------GCAUGUGUCUGCUGC-----------AAAAUAUUAAAGAUUCU------GGAAACCAUAAUCUUAUUGACGUGAUAGGAUUACACUUUGCUAAGUG ---------(((.((((...((.(-----------(........((((((.(------((...))).)))))).......))...))...))))..)))...... ( -14.26, z-score = -0.67, R) >droSec1.super_16 1856379 79 - 1878335 ---------GCAUGUGUCUGCUGC-----------AAAAUAUUAAAGAUUCU------GGAAACCAUAAUCUUAUUGACGUGAUAGGAUUACACUUUGCUAAGUG ---------(((.((((...((.(-----------(........((((((.(------((...))).)))))).......))...))...))))..)))...... ( -14.26, z-score = -0.67, R) >droYak2.chr2L 10140173 90 - 22324452 ---------GCAUGUGUCUGCUGCAAGCAGAAGGAAAAAUAUUAAAGAUUCU------GGAAACCAUAAUCUUAUUGACGUGAUAGGAUUACACUUUGCUAAGUG ---------.(((((.(((((.....))))).............((((((.(------((...))).))))))....))))).........(((((....))))) ( -19.90, z-score = -1.33, R) >droAna3.scaffold_12943 1788801 82 - 5039921 -----------------UCUCUGCUGGCAACAAGAAAAAUAUUAAAGAUUCC------GAAAACCAUAAUCUUAUUGACGUGAUAGGAUUACACUUUGCUAAGUG -----------------..(((..((....))))).................------........((((((((((.....))))))))))(((((....))))) ( -13.00, z-score = -1.02, R) >droEre2.scaffold_4929 14903389 90 + 26641161 ---------GCAUGUGUCUGCUGCAAGCAGCAGGAAAAAUAUUAAAGAUUCU------GGAAACCAUAAUCUUAUUGACGUGAUAGGAUUACACUUUGCUAAGUG ---------.(((((.(((((((....)))))))..........((((((.(------((...))).))))))....))))).........(((((....))))) ( -23.90, z-score = -2.29, R) >droPer1.super_5 3462202 96 - 6813705 ---GAUGGAGAUGGGUGCCCUGGCUGGCAAGAGGAAAAAUAUUAAAGAUUCU------GAAAACCAUAAUCUUAUUGACGUGAUAGGAUUACACUUUGCUAAGUG ---.......((((.((((......))))((((...............))))------.....))))(((((((((.....))))))))).(((((....))))) ( -17.86, z-score = -0.02, R) >droWil1.scaffold_180772 8326871 101 + 8906247 GGCGAGGACAGAGGCGUUGUGCUUGGGUAACAAGAAAAAUAUUAAAGAUUCUCG----AAAAACCAUAAUCUUAUUGACGUGAUAGGAUUACACUUUGCUAAGUG (((((((...((((.(((...((((.....))))............))))))).----........((((((((((.....))))))))))..)))))))..... ( -20.36, z-score = -0.73, R) >droVir3.scaffold_12723 724765 91 + 5802038 --AAACAAGUG------GGCGACUUGGCAACAAGAAAAAUAUUAAAGAUUCUC------AAAACCAUAAUCUUAUUGACGUGAUAGGAUUACACUUUGCUAAGUG --......(((------(....((((....))))............(.....)------....))))(((((((((.....))))))))).(((((....))))) ( -18.20, z-score = -2.05, R) >droMoj3.scaffold_6500 16629973 82 - 32352404 -----------------------GCGACAACAAGAAAAAUAUUAAAGAUUCUCUCUCUUAAAACCAUAAUCCUAUUGACGUGAUAGGAUUACACUUUGCUAAGUG -----------------------.....................((((.......)))).......((((((((((.....))))))))))(((((....))))) ( -13.50, z-score = -2.03, R) >droGri2.scaffold_15126 307241 97 + 8399593 --AAACAAGUGGUGGGCGCUGUCCUGGCAACAAGAAAAAUAUUAAAGAUUCUC------AAAACCAUAAUCUUAUUGACGUGAUAGGAUUACACUUUGCUAAGUG --......(((((((((...))))((....)).....................------...)))))(((((((((.....))))))))).(((((....))))) ( -19.30, z-score = -0.75, R) >consensus _________GCAUGUGUCUGCUGCUGGCAACAAGAAAAAUAUUAAAGAUUCU______GAAAACCAUAAUCUUAUUGACGUGAUAGGAUUACACUUUGCUAAGUG ..................................................................((((((((((.....))))))))))(((((....))))) (-10.83 = -10.75 + -0.08)

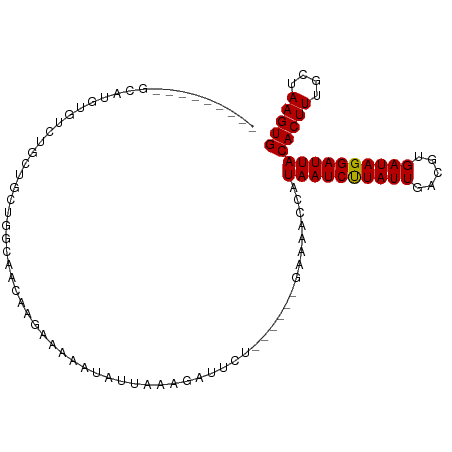
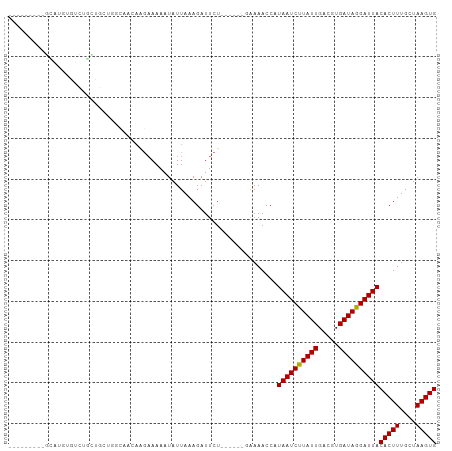
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:38:43 2011