
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 13,728,315 – 13,728,432 |
| Length | 117 |
| Max. P | 0.513686 |

| Location | 13,728,315 – 13,728,432 |
|---|---|
| Length | 117 |
| Sequences | 11 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 62.70 |
| Shannon entropy | 0.81158 |
| G+C content | 0.48118 |
| Mean single sequence MFE | -32.30 |
| Consensus MFE | -7.87 |
| Energy contribution | -7.75 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.55 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.24 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.513686 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 13728315 117 + 23011544 GCCAAAUUAAGUGCUCCCUGGGGUCCAAUCAGCUGGA-GCUCUCCAACUGUUGGACAUUUUGAUGUAUAGUCAUCAAAAUUGCGGCCUGUCGCCCAAAAGCACAGCAAGUCAUCACAA ..........(((((...(((.((((((.(((.((((-....)))).)))))))))(((((((((......))))))))).((((....)))))))..)))))............... ( -39.00, z-score = -3.14, R) >droSim1.chr2L 13486899 117 + 22036055 GCCAAAUUAAGUGCUCCCUGGGGUCCAAUCAGCUGGA-GCUCGCCAACUGUUGGACAUUUUGAUGUAUAGUCGUCAAAAUUGCGGCCUGUCGCCCAAAAGCACAGCAAGUCAUCACAA ..........(((((...(((.((((((.(((.(((.-.....))).)))))))))(((((((((......))))))))).((((....)))))))..)))))............... ( -36.50, z-score = -1.84, R) >droSec1.super_16 1855729 117 + 1878335 GCCAAAUUAAGUGCUCCCUGGGGUCCAAUCAGCUGGA-GCUCGCCAACUGUUGGACAUUUUGAUGUAUAGUCGUCAAAAUUGCGGCCUGUCGCCCAAAAGCACAGCAAGUCAUCACAA ..........(((((...(((.((((((.(((.(((.-.....))).)))))))))(((((((((......))))))))).((((....)))))))..)))))............... ( -36.50, z-score = -1.84, R) >droYak2.chr2L 10139477 117 + 22324452 ACCAAAUUAAGUGCUCCCCGGGGUCCAAUCAGCUGGA-GCUCGCCAACUGUUGGACAUUUUGAUGUAUAGUCGUCAAAAUUGCGGCCCGUCCCCCAAAAGCACAGCAAGUCAUCACAA ..........(((((....(((((((((.(((.(((.-.....))).)))))))))(((((((((......)))))))))...........)))....)))))............... ( -34.80, z-score = -2.22, R) >droAna3.scaffold_12943 1788052 104 + 5039921 ACAGAAUUAAGUGCCCCCUGGGGUCCACUCACCGAAAAGCCCGGCAACUGUUGGACAUUUUGAUGUAUAGCCGUCAAAAUUGC------UCGCCC--------AGCAAGUCAUCACAA ((((......(..(((....)))..).....(((.......)))...))))..(((.((((((((......))))))))((((------(.....--------))))))))....... ( -26.10, z-score = -0.54, R) >droEre2.scaffold_4929 14902724 117 - 26641161 ACCAAAUUAAGUGCUCCCUGGGGUCCAAUCAGCUGGA-GCUCGCCAACUGUUGGACAUUUUGAUGUAUAGUCGUCAAAAUUGCGGCCUGUCCCCCAAAAGCACAGCAAGUCAUCACAA ..........(((((...((((((((((.(((.(((.-.....))).)))))))))(((((((((......)))))))))............))))..)))))............... ( -36.20, z-score = -2.30, R) >dp4.chr4_group1 3533916 102 - 5278887 -GACAAUUAAAUGCUCCCUGGGGUGCCAUCGGCACA---------------UGGACACUUUGAUGAAUAGCCAUCAAAAUUGUUGGCUGUCCCCUGGGGUGCCAGGGGGUCAUCACAA -.........(((((((((((.(((((...))))).---------------....((((..(..(.(((((((.((....)).))))))).)..)..)))))))))))).)))..... ( -39.30, z-score = -1.61, R) >droPer1.super_5 3461520 102 + 6813705 -GACAAUUAAAUGCUCCCUGGGGUGCCAUCGGCACA---------------UGGAGACUUUGAUGAAUAGCCAUCAAAAUUGUUGGCUGUCCCCUGGGGUGCCAGGGGGUGCCCACAA -(((((((.....((((.....(((((...))))).---------------.))))...((((((......)))))))))))))((.(..(((((((....)))))))..).)).... ( -40.90, z-score = -1.43, R) >droWil1.scaffold_180772 8326272 93 - 8906247 GCAACAUUAAAUGCUCCCUGGGGGGCACUCAGCGCUA-----AACAACUGUUGGACAUUUUGAUAUAUAGUUGUCAAAGGAAC-GACGCUCCUCCACAA------------------- ((.........((((((....))))))..........-----.......((((..(.((((((((......)))))))))..)-)))))..........------------------- ( -22.40, z-score = -0.86, R) >droVir3.scaffold_12723 724138 98 - 5802038 CCAACUCCAAGUGCUCCCUGGGGCGCAAUUCAAGCUU-----AAUGAUUGCUGAACAUUUUGCUGGAUAUAUAU-AUUUUUUU-----UUUGUUUGGGGCCUGUCUCAA--------- ....((((((((((((....)))))).(((((.((..-----((((.........))))..)))))))......-........-----.....))))))..........--------- ( -23.50, z-score = -0.46, R) >droGri2.scaffold_15126 306573 97 - 8399593 GCAACUUGAACUGCUCCCUGGGGUGCUAUUCAAGCUA-----AUCGAUUGCUUGAUAUUUUAAUCCAUGGGAAUUACAUUUUU-----AUUGGUCGUGACGUCUCUA----------- (((........)))((((((((.......((((((..-----.......))))))........)))).)))).(((((((...-----...))).))))........----------- ( -20.06, z-score = -0.54, R) >consensus GCAAAAUUAAGUGCUCCCUGGGGUCCAAUCAGCUCGA_GC_CGCCAACUGUUGGACAUUUUGAUGUAUAGUCGUCAAAAUUGCGGCCUGUCGCCCAAAAGCACAGCAAGUCAUCACAA ............((((....))))..................................(((((((......)))))))........................................ ( -7.87 = -7.75 + -0.12)
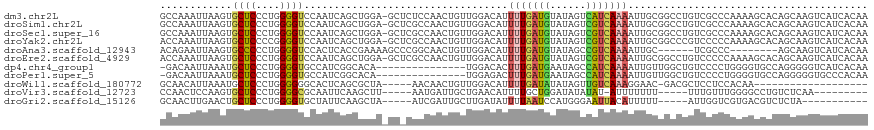

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:38:41 2011