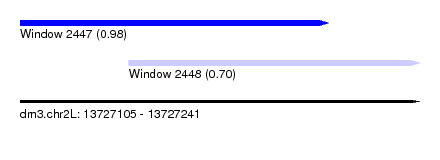
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 13,727,105 – 13,727,241 |
| Length | 136 |
| Max. P | 0.981213 |

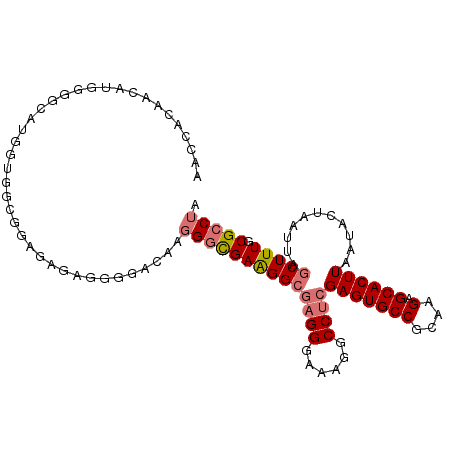
| Location | 13,727,105 – 13,727,210 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 77.30 |
| Shannon entropy | 0.37519 |
| G+C content | 0.53666 |
| Mean single sequence MFE | -35.75 |
| Consensus MFE | -24.02 |
| Energy contribution | -24.90 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.07 |
| SVM RNA-class probability | 0.981213 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 13727105 105 + 23011544 AACCACAACAUGGGGCACGUUGGCGGAGAGAGGAAACGAUGGCGAAGGCGAGGUAAAGGCCUCGAGUGCCGCAAGAGCACUUAAUACUAAUUAGCUUUGCGCCUA ..((.((((.((...)).))))..)).....(....)...((((((((((((((....)))))(((((((....).))))))...........))))).)))).. ( -37.70, z-score = -2.63, R) >droSim1.chr2L 13485659 105 + 22036055 AACCACAACAUGGGGCAUGGUGGUGGAGAGAGGUGACGAGGGCGAAGGCACGGGAAAGGCCUCGAGUGCCGCAAGAGCACUUAAUACUAAUUAGCUUUGCGCCUA ..((((..((((...))))...)))).....(....)..((((((((((.((((......))))((((((....).)))))............))))).))))). ( -37.60, z-score = -2.24, R) >droSec1.super_16 1854488 105 + 1878335 AACCACAACAUGGGGCAUAGUGGUGCAGAGAGGGGACAAGGGCGAAGGCGAGGGAAAGGCCUCGAGUGCCGCAAGAGCACUUAAUACUAAUUAGCUUUGCGCCUA ..(((.....))).((((....)))).....(....)..((((((((((((((......))))(((((((....).))))))...........))))).))))). ( -38.10, z-score = -2.65, R) >droEre2.scaffold_4929 14901540 83 - 26641161 ---------------------AGCCACAACAUGGGGCAUGGUUGAGGGAGAGG-AAUGGCCUCGAGUGCCGCAAGAGCACUUAAUACUAAUUAGCUUUGCGCCUA ---------------------...........((.(((.((((((....((((-.....))))(((((((....).))))))........)))))).))).)).. ( -29.60, z-score = -1.96, R) >consensus AACCACAACAUGGGGCAUGGUGGCGGAGAGAGGGGACAAGGGCGAAGGCGAGGGAAAGGCCUCGAGUGCCGCAAGAGCACUUAAUACUAAUUAGCUUUGCGCCUA .......................................((((((((((((((......))))(((((((....).))))))...........))))).))))). (-24.02 = -24.90 + 0.88)

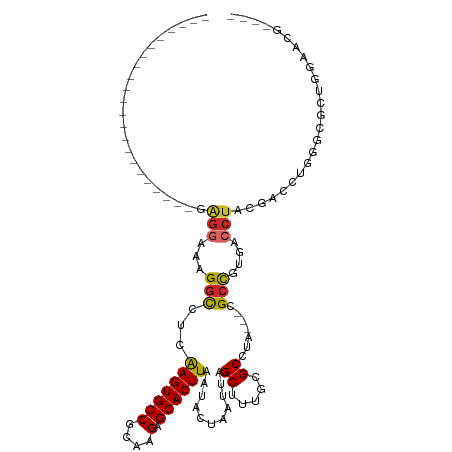
| Location | 13,727,142 – 13,727,241 |
|---|---|
| Length | 99 |
| Sequences | 9 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 72.71 |
| Shannon entropy | 0.50682 |
| G+C content | 0.55930 |
| Mean single sequence MFE | -31.37 |
| Consensus MFE | -14.79 |
| Energy contribution | -14.71 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.701223 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 13727142 99 + 23011544 ---------GAUGGCGAAGGCGAGGUAAAGGCCUCGAGUGCCGCAAGAGCACUUAAUACUAAUUAGCUUUGCGCCUA---CGCCGUGACCUACGACCUGGGCGCUGGAACG---- ---------...((((.(((((((((....)))))(((((((....).))))))..................)))).---))))(((.((((.....))))))).......---- ( -39.90, z-score = -2.92, R) >droSim1.chr2L 13485696 99 + 22036055 ---------GAGGGCGAAGGCACGGGAAAGGCCUCGAGUGCCGCAAGAGCACUUAAUACUAAUUAGCUUUGCGCCUA---CGCCGUGACCUACGACCUGGGCGCUGGAACG---- ---------.(((((((.(((.(......))))))(((((((....).))))))...........)))))(((((((---.(.((((...)))).).))))))).......---- ( -35.60, z-score = -1.20, R) >droSec1.super_16 1854525 99 + 1878335 ---------AAGGGCGAAGGCGAGGGAAAGGCCUCGAGUGCCGCAAGAGCACUUAAUACUAAUUAGCUUUGCGCCUA---CGCCGUGACCUACGACCUGGGCGCUGGAACG---- ---------..((((((((((((((......))))(((((((....).))))))...........))))).))))).---..(((((.((((.....))))))).))....---- ( -38.80, z-score = -2.03, R) >droAna3.scaffold_12943 1786747 86 + 5039921 ----------------------GAGGAAAGGCCUCAAGUGCCGCAAGAGCACUUAAUACUAAUUAGCUUUGCGCCUA---CGACCUGCCCGGACCGUAGCGUAGCAGAGCG---- ----------------------((((.....))))(((((((....).))))))...........((((((((((((---((.((.....))..))))).)).))))))).---- ( -34.80, z-score = -3.91, R) >droEre2.scaffold_4929 14901568 86 - 26641161 ----------------------GAGGAAUGGCCUCGAGUGCCGCAAGAGCACUUAAUACUAAUUAGCUUUGCGCCUA---CGCUGUGACCUACGACCCGGGCGCAGGAACG---- ----------------------((((.....))))(((((((....).)))))).............(((((((((.---.(.((((...)))).)..)))))))))....---- ( -29.20, z-score = -1.87, R) >dp4.chr4_group1 3532681 90 - 5278887 ----------------------GAGGGAAGGCCUCAAGUGCCGCAAGAGCACUUAAUACUAAUUAGCUUUGCGCCUA---CGCCGUGACCUACGACCUGCCCUGGCAAGCGGGUG ----------------------.(((...(((...(((((((....).))))))...........((.....))...---.)))....)))...((((((........)))))). ( -27.60, z-score = 0.04, R) >droPer1.super_5 3460279 90 + 6813705 ----------------------GAGGGAAGGCCUCAAGUGCCGCAAGAGCACUUAAUACUAAUUAGCUUUGCGCCUA---CGCCGUGACCUACGACCUGCCCUGGCAAGCGGGUG ----------------------.(((...(((...(((((((....).))))))...........((.....))...---.)))....)))...((((((........)))))). ( -27.60, z-score = 0.04, R) >droWil1.scaffold_180772 8324728 84 - 8906247 ----------------------AAGAGACGGCCUGAAGUGCCACAAGAGCACUUAAUACUAAUUAGCUUUGCGCCUAUGCCGCCGUGACCUACGACCUG--CUAUGGA------- ----------------------.....(((((...(((((((....).))))))...........((.....)).......))))).........((..--....)).------- ( -16.80, z-score = 0.22, R) >droGri2.scaffold_15126 300300 104 - 8399593 AACAACAACGUCAAAAAAACGUUGAGGGUGGUUUCAAGUGCCGAAAGAGCACUUAAUACUAAUUUGCUUUGCGCCUA---CGCCGUGACCUACGACCUGGCGACCCA-------- .....((((((.......)))))).(((((((...(((((((....).))))))...)))...........((((..---.(.((((...)))).)..)))))))).-------- ( -32.00, z-score = -2.59, R) >consensus ______________________GAGGAAAGGCCUCAAGUGCCGCAAGAGCACUUAAUACUAAUUAGCUUUGCGCCUA___CGCCGUGACCUACGACCUGGGCGCUGGAACG____ .......................(((...(((...(((((((....).))))))...........((.....)).......)))....)))........................ (-14.79 = -14.71 + -0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:38:40 2011