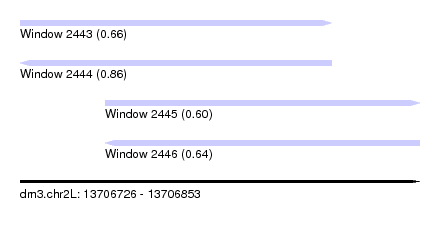
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 13,706,726 – 13,706,853 |
| Length | 127 |
| Max. P | 0.860589 |

| Location | 13,706,726 – 13,706,825 |
|---|---|
| Length | 99 |
| Sequences | 11 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 71.32 |
| Shannon entropy | 0.57608 |
| G+C content | 0.54775 |
| Mean single sequence MFE | -31.04 |
| Consensus MFE | -17.30 |
| Energy contribution | -17.26 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.44 |
| Mean z-score | -0.81 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.661450 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
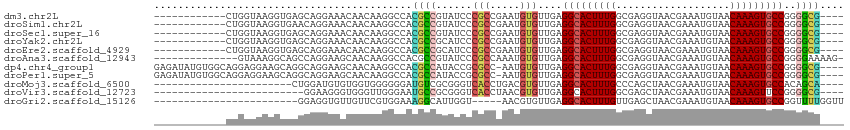
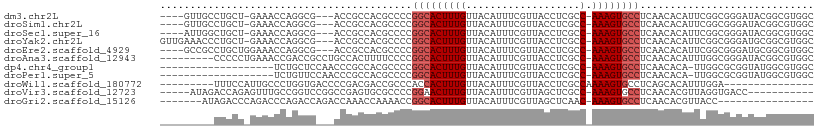
>dm3.chr2L 13706726 99 + 23011544 ------------CUGGUAAGGUGAGCAGGAAACAACAAGGCCACGCCGUAUCCCGCCGAAUGUGUUGAGGCACUUUGGCGAGGUAACGAAAUGUAACAAAGUGCCGGGGCG---- ------------((((((...((....(....).(((.(((...)))((..(((((((((.((((....))))))))))).))..))....)))..))...))))))....---- ( -32.60, z-score = -0.88, R) >droSim1.chr2L 13465373 99 + 22036055 ------------CUGGUAAGGUGAACAGGAAACAACAAGGCCACGCCGUAUCCCGCCGAAUGUGUUGAGGCACUUUGGCGAGGUAACGAAAUGUAACAAAGUGCCGGGGCG---- ------------((((((...((....(....).(((.(((...)))((..(((((((((.((((....))))))))))).))..))....)))..))...))))))....---- ( -32.60, z-score = -1.18, R) >droSec1.super_16 1834192 99 + 1878335 ------------CUGGUAAGGUGAGCAGGAAACAACAAGGCCACGCCGUAUCCCGCCGAAUGUGUUGAGGCACUUUGGCGAGGUAACGAAAUGUAACAAAGUGCCGGGGCG---- ------------((((((...((....(....).(((.(((...)))((..(((((((((.((((....))))))))))).))..))....)))..))...))))))....---- ( -32.60, z-score = -0.88, R) >droYak2.chr2L 10117583 99 + 22324452 ------------CUGGUAAGGUGAGCAGGAAACAACAAGGCCACGCCGCAUCCCGCCGAAUGUGUUGAGGCACUUUGGCGAGGUAACGAAAUGUAACAAAGUGCCGGGGCG---- ------------((((((...((.((.(....).....(((...)))))..(((((((((.((((....))))))))))).)).............))...))))))....---- ( -33.10, z-score = -0.84, R) >droEre2.scaffold_4929 14880098 99 - 26641161 ------------CUGGUAAGGUGAGCAGGAAACAACAAGGCCACGCCGCAUCCCGCCGAAUGUGUUGAGGCACUUUGGCGAGGUAACGAAAUGUAACAAAGUGCCGGGGCG---- ------------((((((...((.((.(....).....(((...)))))..(((((((((.((((....))))))))))).)).............))...))))))....---- ( -33.10, z-score = -0.84, R) >droAna3.scaffold_12943 1762135 100 + 5039921 --------------GUAAAGGCAGCCAGGAAGCAACAAGGCCACGCCGUAUCCCGCCAAAUGUGUUGAGGCACUUUGGCGAGGUAACGAAAUGUAACAAAGUGCCGGGGAAAAG- --------------.....(((.(((..(......)..)))...)))...((((((((((.((((....))))))))))..((((.(.............))))).))))....- ( -29.92, z-score = -0.79, R) >dp4.chr4_group1 3502899 110 - 5278887 GAGAUAUGUGGCAGGAGGAAGCAGGCAGGAAGCAACAAGGCCACGCCAUACCGCGCC-AAUGUGUUGAGGCACUUUGGCGAGGUAACGAAAUGUAACAAAGUGCCGGGGCG---- .......(((((....(....)..((.....))......)))))(((.((((.((((-((.((((....)))).)))))).)))).((..((........))..)).))).---- ( -36.10, z-score = -1.39, R) >droPer1.super_5 3430441 110 + 6813705 GAGAUAUGUGGCAGGAGGAAGCAGGCAGGAAGCAACAAGGCCACGCCAUACCGCGCC-AAUGUGUUGAGGCACUUUGGCGAGGUAACGAAAUGUAACAAAGUGCCGGGGCG---- .......(((((....(....)..((.....))......)))))(((.((((.((((-((.((((....)))).)))))).)))).((..((........))..)).))).---- ( -36.10, z-score = -1.39, R) >droMoj3.scaffold_6500 16588897 88 + 32352404 -----------------------CUGGAUGUGUGGUGGGGGGAUGUCGCGGGUCACCUGACGUGUUGAGGCACUUUGCCCAGCUAACGAAAUGUAACAAAGUGCCACAGCA---- -----------------------...(((.((((..(......)..)))).)))........(((((.(((((((((..((.(....)...))...))))))))).)))))---- ( -28.90, z-score = -0.06, R) >droVir3.scaffold_12723 687577 86 - 5802038 -------------------------GGAAGGGUGGGUUGGGAAUGCCGCGGGUCACCUAACGUGUUGAGGCACUUUGGCGAGCUAACGAAAUGUAACAAAGUUCCGGGGCG---- -------------------------....((((((.(((((....)).))).)))))).....(((..((.(((((((((..(....)...)))..)))))).))..))).---- ( -23.70, z-score = 0.54, R) >droGri2.scaffold_15126 262933 86 - 8399593 ------------------------GGAGGUGUUGUUCGUGGAAAGGCAUUGGU-----AACGUGUUGAGGCACUUUGUUGAGCUAACGAAAUGUAACAAAGUGCCGGUUUUGGUU ------------------------...(((((..(((...)))..)))))...-----..........((((((((((((..(....).....)))))))))))).......... ( -22.70, z-score = -1.25, R) >consensus ____________CUGGUAAGGUGAGCAGGAAACAACAAGGCCACGCCGUAUCCCGCCGAAUGUGUUGAGGCACUUUGGCGAGGUAACGAAAUGUAACAAAGUGCCGGGGCG____ ...........................................((((......(((.....)))....(((((((((...................)))))))))..)))).... (-17.30 = -17.26 + -0.05)
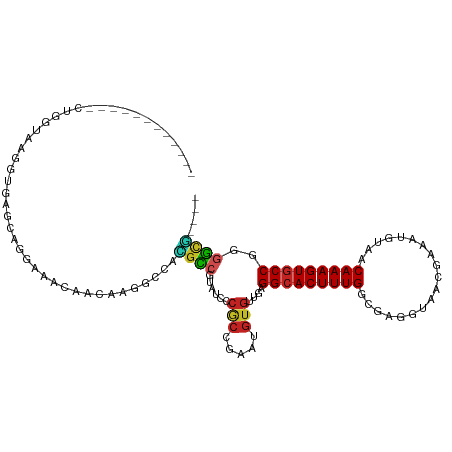
| Location | 13,706,726 – 13,706,825 |
|---|---|
| Length | 99 |
| Sequences | 11 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 71.32 |
| Shannon entropy | 0.57608 |
| G+C content | 0.54775 |
| Mean single sequence MFE | -26.70 |
| Consensus MFE | -12.63 |
| Energy contribution | -12.82 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.860589 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 13706726 99 - 23011544 ----CGCCCCGGCACUUUGUUACAUUUCGUUACCUCGCCAAAGUGCCUCAACACAUUCGGCGGGAUACGGCGUGGCCUUGUUGUUUCCUGCUCACCUUACCAG------------ ----......(((((((((...................)))))))))...........(((((((.((((((......)))))).)))))))...........------------ ( -29.11, z-score = -1.67, R) >droSim1.chr2L 13465373 99 - 22036055 ----CGCCCCGGCACUUUGUUACAUUUCGUUACCUCGCCAAAGUGCCUCAACACAUUCGGCGGGAUACGGCGUGGCCUUGUUGUUUCCUGUUCACCUUACCAG------------ ----......(((((((((...................)))))))))...........(((((((.((((((......)))))).)))))))...........------------ ( -26.61, z-score = -0.99, R) >droSec1.super_16 1834192 99 - 1878335 ----CGCCCCGGCACUUUGUUACAUUUCGUUACCUCGCCAAAGUGCCUCAACACAUUCGGCGGGAUACGGCGUGGCCUUGUUGUUUCCUGCUCACCUUACCAG------------ ----......(((((((((...................)))))))))...........(((((((.((((((......)))))).)))))))...........------------ ( -29.11, z-score = -1.67, R) >droYak2.chr2L 10117583 99 - 22324452 ----CGCCCCGGCACUUUGUUACAUUUCGUUACCUCGCCAAAGUGCCUCAACACAUUCGGCGGGAUGCGGCGUGGCCUUGUUGUUUCCUGCUCACCUUACCAG------------ ----......(((((((((...................)))))))))...........(((((((.((((((......)))))).)))))))...........------------ ( -29.21, z-score = -1.19, R) >droEre2.scaffold_4929 14880098 99 + 26641161 ----CGCCCCGGCACUUUGUUACAUUUCGUUACCUCGCCAAAGUGCCUCAACACAUUCGGCGGGAUGCGGCGUGGCCUUGUUGUUUCCUGCUCACCUUACCAG------------ ----......(((((((((...................)))))))))...........(((((((.((((((......)))))).)))))))...........------------ ( -29.21, z-score = -1.19, R) >droAna3.scaffold_12943 1762135 100 - 5039921 -CUUUUCCCCGGCACUUUGUUACAUUUCGUUACCUCGCCAAAGUGCCUCAACACAUUUGGCGGGAUACGGCGUGGCCUUGUUGCUUCCUGGCUGCCUUUAC-------------- -.........((((........(((.((((...((((((((((((......))).)))))))))..)))).)))(((..(......)..))))))).....-------------- ( -31.60, z-score = -2.23, R) >dp4.chr4_group1 3502899 110 + 5278887 ----CGCCCCGGCACUUUGUUACAUUUCGUUACCUCGCCAAAGUGCCUCAACACAUU-GGCGCGGUAUGGCGUGGCCUUGUUGCUUCCUGCCUGCUUCCUCCUGCCACAUAUCUC ----.(((..(((.....))).........((((.((((((.(((......))).))-)))).)))).)))(((((......((.........))........)))))....... ( -31.54, z-score = -1.78, R) >droPer1.super_5 3430441 110 - 6813705 ----CGCCCCGGCACUUUGUUACAUUUCGUUACCUCGCCAAAGUGCCUCAACACAUU-GGCGCGGUAUGGCGUGGCCUUGUUGCUUCCUGCCUGCUUCCUCCUGCCACAUAUCUC ----.(((..(((.....))).........((((.((((((.(((......))).))-)))).)))).)))(((((......((.........))........)))))....... ( -31.54, z-score = -1.78, R) >droMoj3.scaffold_6500 16588897 88 - 32352404 ----UGCUGUGGCACUUUGUUACAUUUCGUUAGCUGGGCAAAGUGCCUCAACACGUCAGGUGACCCGCGACAUCCCCCCACCACACAUCCAG----------------------- ----((.((.(((((((((((...............))))))))))).)).)).(((.((....))..))).....................----------------------- ( -21.76, z-score = -0.20, R) >droVir3.scaffold_12723 687577 86 + 5802038 ----CGCCCCGGAACUUUGUUACAUUUCGUUAGCUCGCCAAAGUGCCUCAACACGUUAGGUGACCCGCGGCAUUCCCAACCCACCCUUCC------------------------- ----.(((.(((......((((........))))(((((...(((......)))....))))).))).)))...................------------------------- ( -19.00, z-score = -1.10, R) >droGri2.scaffold_15126 262933 86 + 8399593 AACCAAAACCGGCACUUUGUUACAUUUCGUUAGCUCAACAAAGUGCCUCAACACGUU-----ACCAAUGCCUUUCCACGAACAACACCUCC------------------------ ..........(((((((((((...............)))))))))))..........-----.............................------------------------ ( -14.96, z-score = -2.64, R) >consensus ____CGCCCCGGCACUUUGUUACAUUUCGUUACCUCGCCAAAGUGCCUCAACACAUUCGGCGGGAUACGGCGUGGCCUUGUUGCUUCCUGCUCACCUUACCAG____________ ....((((..(((((((((...................)))))))))...........(........)))))........................................... (-12.63 = -12.82 + 0.19)
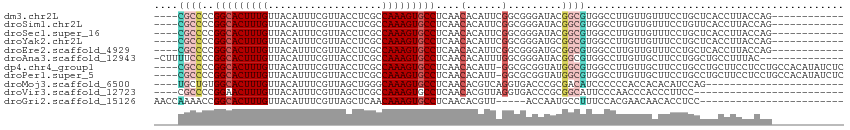
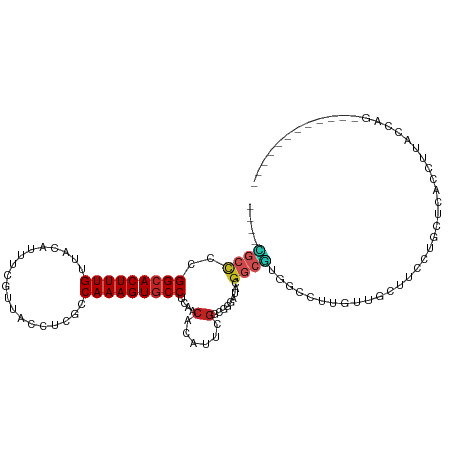
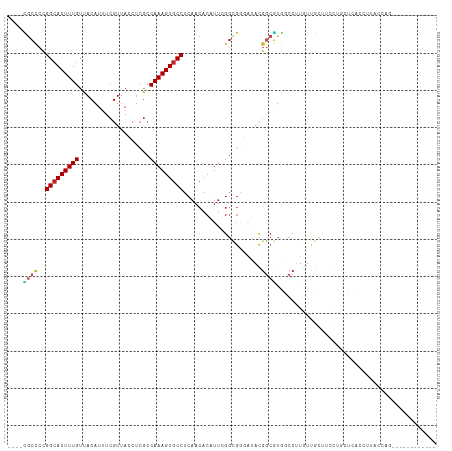
| Location | 13,706,753 – 13,706,853 |
|---|---|
| Length | 100 |
| Sequences | 11 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 69.03 |
| Shannon entropy | 0.67132 |
| G+C content | 0.57458 |
| Mean single sequence MFE | -37.14 |
| Consensus MFE | -11.12 |
| Energy contribution | -11.64 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.30 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.602802 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 13706753 100 + 23011544 GCCACGCCGUAUCCCGCCGAAUGUGUUGAGGCACUUU-GGCGAGGUAACGAAAUGUAACAAAGUGCCGGGGCGUGGCGGU---CGCCUGGUUUC-AGCAGGCAAC---- ((((((((.((((.(((((((.((((....)))))))-)))).)))).((..((........))..)).))))))))...---.(((((.....-..)))))...---- ( -44.50, z-score = -2.47, R) >droSim1.chr2L 13465400 100 + 22036055 GCCACGCCGUAUCCCGCCGAAUGUGUUGAGGCACUUU-GGCGAGGUAACGAAAUGUAACAAAGUGCCGGGGCGUGGCGGU---CGCCUGGUUUC-AGCAGGCAAC---- ((((((((.((((.(((((((.((((....)))))))-)))).)))).((..((........))..)).))))))))...---.(((((.....-..)))))...---- ( -44.50, z-score = -2.47, R) >droSec1.super_16 1834219 100 + 1878335 GCCACGCCGUAUCCCGCCGAAUGUGUUGAGGCACUUU-GGCGAGGUAACGAAAUGUAACAAAGUGCCGGGGCGUGGCGGU---CGCCUGGUUUC-AGCAGCCAAU---- ((((((((.((((.(((((((.((((....)))))))-)))).)))).((..((........))..)).))))))))(((---.((.((....)-))).)))...---- ( -42.20, z-score = -1.90, R) >droYak2.chr2L 10117610 104 + 22324452 GCCACGCCGCAUCCCGCCGAAUGUGUUGAGGCACUUU-GGCGAGGUAACGAAAUGUAACAAAGUGCCGGGGCGUGGCGGU---CGCCUGGUUUC-AGCAGGGUUUCAAC ((((((((.(....(((((((.((((....)))))))-)))).((((.(.............)))))).)))))))).(.---.((((.((...-.)).))))..)... ( -42.52, z-score = -1.46, R) >droEre2.scaffold_4929 14880125 101 - 26641161 GCCACGCCGCAUCCCGCCGAAUGUGUUGAGGCACUUU-GGCGAGGUAACGAAAUGUAACAAAGUGCCGGGGCGUGGCGGU---CGCCUGGUUUCCAGCAGGCGGC---- ((((((((.(....(((((((.((((....)))))))-)))).((((.(.............)))))).)))))))).((---((((((........))))))))---- ( -50.72, z-score = -3.02, R) >droAna3.scaffold_12943 1762160 99 + 5039921 GCCACGCCGUAUCCCGCCAAAUGUGUUGAGGCACUUU-GGCGAGGUAACGAAAUGUAACAAAGUGCCGGGGAAAAGUGGCAGGCGGUCGGUUUCAGGGGG--------- (((.((((((..(((((((((.((((....)))))))-)))).))..))..............(((((........)))))))))...))).........--------- ( -33.60, z-score = -0.44, R) >dp4.chr4_group1 3502938 88 - 5278887 GCCACGCCAUACCGCGCCAA-UGUGUUGAGGCACUUU-GGCGAGGUAACGAAAUGUAACAAAGUGCCGGGGCGUGGCGGGUUGGAGCAGA------------------- ((((((((.((((.((((((-.((((....)))).))-)))).)))).((..((........))..)).)))))))).............------------------- ( -39.20, z-score = -2.70, R) >droPer1.super_5 3430480 88 + 6813705 GCCACGCCAUACCGCGCCAA-UGUGUUGAGGCACUUU-GGCGAGGUAACGAAAUGUAACAAAGUGCCGGGGCGUGGCGGGUUGGAACAGA------------------- ((((((((.((((.((((((-.((((....)))).))-)))).)))).((..((........))..)).)))))))).............------------------- ( -39.20, z-score = -3.01, R) >droWil1.scaffold_180772 8298026 85 - 8906247 ---------------UCCAAAUGUGCUGAGGCACUUUUGGCGAGGUAACGAAAUGUAACAAAGUGGUGGGCGGUCGUCGGGGUCACCAGGGCAAUGGAAA--------- ---------------((((..(((.(((..(.(((((.(((((.....((..((........))..)).....)))))))))))..))).))).))))..--------- ( -20.70, z-score = 0.61, R) >droVir3.scaffold_12723 687602 92 - 5802038 -----------GGUCACCUAACGUGUUGAGGCACUUU-GGCGAGCUAACGAAAUGUAACAAAGUUCCGGGGCGCACUCGGCCGGACCGGCAAACUCUGGUCUAU----- -----------(((((((....).)).(((((.((((-((..((((...............)))))))))).)).)))))))(((((((......)))))))..----- ( -27.66, z-score = -0.13, R) >droGri2.scaffold_15126 262959 85 - 8399593 ----------------GGUAACGUGUUGAGGCACUUU-GUUGAGCUAACGAAAUGUAACAAAGUGCCGGUUUUGGUUUGGUCUGGUCUGGGUCUGGGUCUAU------- ----------------.............((((((((-((((..(....).....))))))))))))...........(..(..(.......)..)..)...------- ( -23.70, z-score = -1.48, R) >consensus GCCACGCCGUAUCCCGCCGAAUGUGUUGAGGCACUUU_GGCGAGGUAACGAAAUGUAACAAAGUGCCGGGGCGUGGCGGG___CGCCUGGUUUC_AGCAG_________ .......................((((..((((((((..(((.(....)....)))...))))))))..)))).................................... (-11.12 = -11.64 + 0.52)
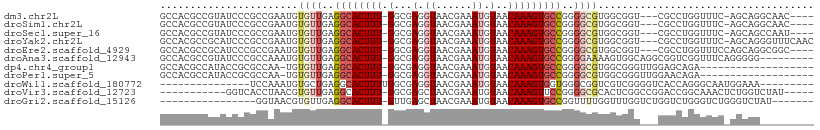

| Location | 13,706,753 – 13,706,853 |
|---|---|
| Length | 100 |
| Sequences | 11 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 69.03 |
| Shannon entropy | 0.67132 |
| G+C content | 0.57458 |
| Mean single sequence MFE | -31.90 |
| Consensus MFE | -7.61 |
| Energy contribution | -7.89 |
| Covariance contribution | 0.27 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.24 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.640419 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 13706753 100 - 23011544 ----GUUGCCUGCU-GAAACCAGGCG---ACCGCCACGCCCCGGCACUUUGUUACAUUUCGUUACCUCGCC-AAAGUGCCUCAACACAUUCGGCGGGAUACGGCGUGGC ----((((((((..-.....))))))---)).((((((((..(((((((((...................)-)))))))).......((((....))))..)))))))) ( -41.01, z-score = -3.03, R) >droSim1.chr2L 13465400 100 - 22036055 ----GUUGCCUGCU-GAAACCAGGCG---ACCGCCACGCCCCGGCACUUUGUUACAUUUCGUUACCUCGCC-AAAGUGCCUCAACACAUUCGGCGGGAUACGGCGUGGC ----((((((((..-.....))))))---)).((((((((..(((((((((...................)-)))))))).......((((....))))..)))))))) ( -41.01, z-score = -3.03, R) >droSec1.super_16 1834219 100 - 1878335 ----AUUGGCUGCU-GAAACCAGGCG---ACCGCCACGCCCCGGCACUUUGUUACAUUUCGUUACCUCGCC-AAAGUGCCUCAACACAUUCGGCGGGAUACGGCGUGGC ----...((.((((-.......))))---.))((((((((..(((((((((...................)-)))))))).......((((....))))..)))))))) ( -35.31, z-score = -1.17, R) >droYak2.chr2L 10117610 104 - 22324452 GUUGAAACCCUGCU-GAAACCAGGCG---ACCGCCACGCCCCGGCACUUUGUUACAUUUCGUUACCUCGCC-AAAGUGCCUCAACACAUUCGGCGGGAUGCGGCGUGGC ((((....((((..-.....))))))---)).((((((((..(((((((((...................)-))))))))......(((((....))))).)))))))) ( -36.51, z-score = -1.25, R) >droEre2.scaffold_4929 14880125 101 + 26641161 ----GCCGCCUGCUGGAAACCAGGCG---ACCGCCACGCCCCGGCACUUUGUUACAUUUCGUUACCUCGCC-AAAGUGCCUCAACACAUUCGGCGGGAUGCGGCGUGGC ----..((((((..(....)))))))---...((((((((..(((((((((...................)-))))))))......(((((....))))).)))))))) ( -43.81, z-score = -2.65, R) >droAna3.scaffold_12943 1762160 99 - 5039921 ---------CCCCCUGAAACCGACCGCCUGCCACUUUUCCCCGGCACUUUGUUACAUUUCGUUACCUCGCC-AAAGUGCCUCAACACAUUUGGCGGGAUACGGCGUGGC ---------..........(((..((((((((..........))))..............((...((((((-((((((......))).)))))))))..))))))))). ( -29.00, z-score = -1.45, R) >dp4.chr4_group1 3502938 88 + 5278887 -------------------UCUGCUCCAACCCGCCACGCCCCGGCACUUUGUUACAUUUCGUUACCUCGCC-AAAGUGCCUCAACACA-UUGGCGCGGUAUGGCGUGGC -------------------.............((((((((..(((.....))).........((((.((((-((.(((......))).-)))))).)))).)))))))) ( -34.40, z-score = -3.43, R) >droPer1.super_5 3430480 88 - 6813705 -------------------UCUGUUCCAACCCGCCACGCCCCGGCACUUUGUUACAUUUCGUUACCUCGCC-AAAGUGCCUCAACACA-UUGGCGCGGUAUGGCGUGGC -------------------.............((((((((..(((.....))).........((((.((((-((.(((......))).-)))))).)))).)))))))) ( -34.40, z-score = -3.55, R) >droWil1.scaffold_180772 8298026 85 + 8906247 ---------UUUCCAUUGCCCUGGUGACCCCGACGACCGCCCACCACUUUGUUACAUUUCGUUACCUCGCCAAAAGUGCCUCAGCACAUUUGGA--------------- ---------..((((......((((((....(((((......((......))......)))))...))))))...((((....))))...))))--------------- ( -18.00, z-score = -1.62, R) >droVir3.scaffold_12723 687602 92 + 5802038 -----AUAGACCAGAGUUUGCCGGUCCGGCCGAGUGCGCCCCGGAACUUUGUUACAUUUCGUUAGCUCGCC-AAAGUGCCUCAACACGUUAGGUGACC----------- -----......((((((...((((...(((.......))))))).))))))...............(((((-...(((......)))....)))))..----------- ( -22.50, z-score = 0.52, R) >droGri2.scaffold_15126 262959 85 + 8399593 -------AUAGACCCAGACCCAGACCAGACCAAACCAAAACCGGCACUUUGUUACAUUUCGUUAGCUCAAC-AAAGUGCCUCAACACGUUACC---------------- -------...................................(((((((((((...............)))-)))))))).............---------------- ( -14.96, z-score = -2.49, R) >consensus _________CUGCU_GAAACCAGGCG___ACCGCCACGCCCCGGCACUUUGUUACAUUUCGUUACCUCGCC_AAAGUGCCUCAACACAUUCGGCGGGAUACGGCGUGGC ..........................................((((((((......................))))))))............................. ( -7.61 = -7.89 + 0.27)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:38:38 2011