
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 13,706,349 – 13,706,463 |
| Length | 114 |
| Max. P | 0.849420 |

| Location | 13,706,349 – 13,706,463 |
|---|---|
| Length | 114 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.72 |
| Shannon entropy | 0.22487 |
| G+C content | 0.41040 |
| Mean single sequence MFE | -29.96 |
| Consensus MFE | -20.28 |
| Energy contribution | -20.23 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.849420 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 13706349 114 - 23011544 ---AAUUCAAGCUCUCUUUUUGUCGCCUUAUUGCCAUUGAAGUUUGUUUUUUAUGGCCUGGCC--GAGCUUGACAAUAAGCACGGCGGCGGC-AAAACUUUUCCACUUCUCAUUUAUUAU ---..............((((((((((.....(((((.((((.....)))).)))))...(((--(.(((((....))))).))))))))))-))))....................... ( -34.30, z-score = -3.53, R) >droSim1.chr2L 13465004 114 - 22036055 ---AAUUCAAGCUCUCUUUUUGUCGCCUUAUUGCCAUUGAAGUUUGUUUUUUAUGGCCUGGCC--GAGCUUGACAAUAAGCACGGCGGCGGC-AAAACUUUUCCACUUCUCAUUUAUUAU ---..............((((((((((.....(((((.((((.....)))).)))))...(((--(.(((((....))))).))))))))))-))))....................... ( -34.30, z-score = -3.53, R) >droSec1.super_16 1833821 114 - 1878335 ---AAUUCAAGCUCUCUUUUUGUCGCCUUAUUGCCAUUGAAGUUUGUUUUUUAUGGCCUGGCC--GAGCUUGACAAUAAGCACGGCGGCGGC-AAAACUUUUCCACUUCUCAUUUAUUAU ---..............((((((((((.....(((((.((((.....)))).)))))...(((--(.(((((....))))).))))))))))-))))....................... ( -34.30, z-score = -3.53, R) >droYak2.chr2L 10117202 114 - 22324452 ---UAUUCAAGCUCUCUUUUUGUCGCCUUAUUGCCAUUGAAGUUUGUUUUUUAUGGCCUGGCC--GAGCUUGACAAUAAGCACGGCGGCGGC-AAAACUUUUCCACUUCUCAUUUAUUAU ---..............((((((((((.....(((((.((((.....)))).)))))...(((--(.(((((....))))).))))))))))-))))....................... ( -34.30, z-score = -3.60, R) >droEre2.scaffold_4929 14879708 114 + 26641161 ---UAUUCAAGCUCUCUUUUUGUCGCCUUAUUGCCAUUGAAGUUUGUUUUUUAUGGCCUGGCC--GAGCUUGACAAUAAGCACGGCGGCGGC-AAAACUUUUCCACUUCUCAUUUAUUAU ---..............((((((((((.....(((((.((((.....)))).)))))...(((--(.(((((....))))).))))))))))-))))....................... ( -34.30, z-score = -3.60, R) >droAna3.scaffold_12943 1761786 112 - 5039921 -----UCCCAGCCCGCUCCUUGUCGCCUUAUUGCUAUUGAAGUUUGUUUUUUAUGGCCUGGCC--GAGCUUGACAAUAAGCACGGCGGCGGC-AAAACUUUUCCACUUCUCAUUUAUUAU -----..............((((((((.....(((((.((((.....)))).)))))...(((--(.(((((....))))).))))))))))-))......................... ( -30.40, z-score = -1.61, R) >dp4.chr4_group1 3502504 107 + 5278887 ---------UGCUC-UUUUUUGUCGCCUUAUUGCUAUUGAAGUUUGUUUUUUAUGGCCUGGCC--GAGCUUGACAAUAAGCACGGCGGCGGC-GAAACUUUUCCACUUCUCAUUUAUUAU ---------.....-..((((((((((.....(((((.((((.....)))).)))))...(((--(.(((((....))))).))))))))))-))))....................... ( -31.70, z-score = -2.90, R) >droPer1.super_5 3430058 107 - 6813705 ---------UGCUC-UUUUUUGUCGCCUUAUUGCUAUUGAAGUUUGUUUUUUAUGGCCUGGCC--GAGCUUGACAAUAAGCACGGCGGCGGC-GAAACUUUUCCACUUCUCAUUUAUUAU ---------.....-..((((((((((.....(((((.((((.....)))).)))))...(((--(.(((((....))))).))))))))))-))))....................... ( -31.70, z-score = -2.90, R) >droWil1.scaffold_180772 8297718 103 + 8906247 ---------------UUUUUCGUCGCCUUAUUGCUAUUGAAGUUUGUUUUUUAUGGCCUGGCC--GAGCUUGACAAUAAGCACGAUGGCGGCAAAAACUUUUCCACUUCUCAUUUAUUAU ---------------......((((((....((((((((((((((.........(((...)))--))))))..)))).))))....))))))............................ ( -21.80, z-score = -0.76, R) >droMoj3.scaffold_6500 16588556 108 - 32352404 -----------CGUCGUUUUUGUCGCCUUAUUGCUAUUGAAGUUUGUUUUUUAUGGCCUGGCCGAGAGCUUGACAAUAAGCACGUUGGCUGC-AAAACUUUUCCACUUCUCAUUUAUUAU -----------......((((((.(((....((((((((...(..((((((...(((...)))))))))..).)))).))))....))).))-))))....................... ( -21.00, z-score = 0.14, R) >droVir3.scaffold_12723 687274 111 + 5802038 ------UUUCUACCAUUUUUUGUCGCCUUAUUGCUAUUGAAGUUUGUUUUUUAUGGCCUGGCC--GAGCUUGACAAUAAGCACGUUGGCGGC-AAAACUUUUCCACUUCUCAUUUAUUAU ------...........((((((((((....((((((((((((((.........(((...)))--))))))..)))).))))....))))))-))))....................... ( -24.80, z-score = -1.76, R) >droGri2.scaffold_15126 262624 117 + 8399593 UGUUUAUUCCUCCCAUUUUUUGUCGCCUUAUUGCUAUUGAAGUUGGUUUUUUAUGGGCUGGCC--GAGCUUGACAAUAAGCACGUUGGCGGC-AAAACUUUUCCACUUCUCAUUUAUUAU .................((((((((((....(((((((((((((((((...........))))--.)))))..)))).))))....))))))-))))....................... ( -26.60, z-score = -0.97, R) >consensus _____UUCAAGCUCUCUUUUUGUCGCCUUAUUGCUAUUGAAGUUUGUUUUUUAUGGCCUGGCC__GAGCUUGACAAUAAGCACGGCGGCGGC_AAAACUUUUCCACUUCUCAUUUAUUAU ................((((.((((((.....(((((.((((.....)))).)))))..........(((((....))))).....)))))).))))....................... (-20.28 = -20.23 + -0.05)

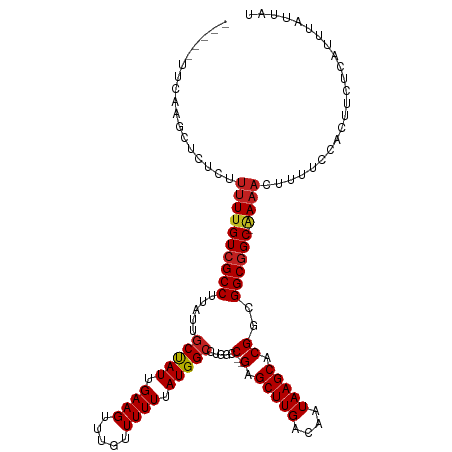

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:38:34 2011