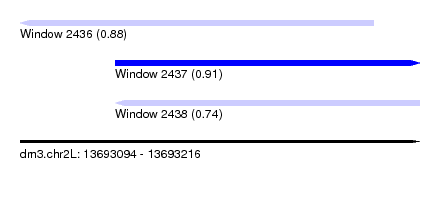
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 13,693,094 – 13,693,216 |
| Length | 122 |
| Max. P | 0.908684 |

| Location | 13,693,094 – 13,693,202 |
|---|---|
| Length | 108 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.14 |
| Shannon entropy | 0.13910 |
| G+C content | 0.32667 |
| Mean single sequence MFE | -17.54 |
| Consensus MFE | -17.10 |
| Energy contribution | -17.06 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.97 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.879317 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
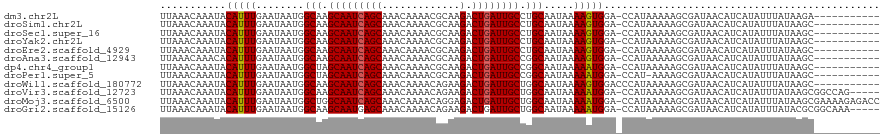
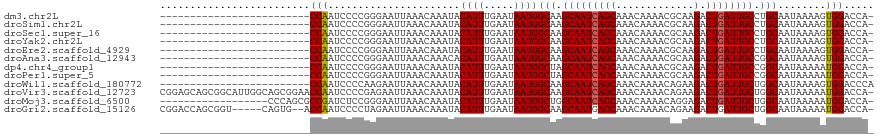
>dm3.chr2L 13693094 108 - 23011544 UUAAACAAAUACAUUUGAAUAAUGGCAAGCAAUCAGCAAACAAAACGCAAGACUGAUUGCCUGCAAUAAAAGUGGA-CCAUAAAAAGCGAUAACAUCAUAUUUAUAAGA----------- ...........(((((........(((.(((((((((.............).)))))))).))).....)))))..-................................----------- ( -16.84, z-score = -1.34, R) >droSim1.chr2L 13451257 108 - 22036055 UUAAACAAAUACAUUUGAAUAAUGGCAAGCAAUCAGCAAACAAAACGCAAGACUGAUUGCCUGCAAUAAAAGUGGA-CCAUAAAAAGCGAUAACAUCAUAUUUAUAAGC----------- ...........(((((........(((.(((((((((.............).)))))))).))).....)))))..-................................----------- ( -16.84, z-score = -1.02, R) >droSec1.super_16 1819498 108 - 1878335 UUAAACAAAUACAUUUGAAUAAUGGCAAGCAAUCAGCAAACAAAACGCAAGACUGAUUGCCUGCAAUAAAAGUGGA-CCAUAAAAAGCGAUAACAUCAUAUUUAUAAGC----------- ...........(((((........(((.(((((((((.............).)))))))).))).....)))))..-................................----------- ( -16.84, z-score = -1.02, R) >droYak2.chr2L 10102568 108 - 22324452 UUAAACAAAUACAUUUGAAUAAUGGCAAGCAAUCAGCAAACAAAACGCAAGACUGAUUGCCUGCAAUAAAAGUGGA-CCAUAAAAAGCGAUAACAUCAUAUUUAUAAGC----------- ...........(((((........(((.(((((((((.............).)))))))).))).....)))))..-................................----------- ( -16.84, z-score = -1.02, R) >droEre2.scaffold_4929 14866611 108 + 26641161 UUAAACAAAUACAUUUGAAUAAUGGCAAGCAAUCAGCAAACAAAACGCAAGACUGAUUGCCUGCAAUAAAAGUGGA-CCAUAAAAAGCGAUAACAUCAUAUUUAUAAGC----------- ...........(((((........(((.(((((((((.............).)))))))).))).....)))))..-................................----------- ( -16.84, z-score = -1.02, R) >droAna3.scaffold_12943 1746931 108 - 5039921 UUAAACAAACACAUUUGAAUAAUGGCAAGCAAUCAGCAAACAAAACGCAAGACUGAUUGCCGGCAAUAAAAGUGGA-CCAUAAAAAGCGAUAACAUCAUAUUUAUAAGC----------- ...........(((((........((..(((((((((.............).))))))))..)).....)))))..-................................----------- ( -15.64, z-score = -0.70, R) >dp4.chr4_group1 3486930 108 + 5278887 UUAAACAAAUACAUUUGAAUAAUGGCUAGCAAUCAGCAAACAAAACGCAAGACUGAUUGCCGGCAAUAAAAAUGGA-CCAUAAAAAGCGAUAACAUCAUAUUUAUAAGC----------- ...........(((((........((..(((((((((.............).))))))))..)).....)))))..-................................----------- ( -17.34, z-score = -1.51, R) >droPer1.super_5 3414805 107 - 6813705 UUAAACAAAUACAUUUGAAUAAUGGCUAGCAAUCAGCAAACAAAACGCAAGACUGAUUGCCGGCAAUAAAAAUGGA-CCAU-AAAAGCGAUAACAUCAUAUUUAUAAGC----------- ...........(((((........((..(((((((((.............).))))))))..)).....)))))..-....-...........................----------- ( -17.34, z-score = -1.58, R) >droWil1.scaffold_180772 627969 109 - 8906247 UUAAACAAAUACAUUUGAAUAAUGGCAAGCAAUCAGCAAACAAAACAGAAGACUGAUUGCUGGCAAUAAAAGUGGACCCAUAAAAAGCGAUAACAUCAUAUUUAUAAGC----------- ...............(((((((((((.((((((((((.............).))))))))).)).......(((....)))............)))..)))))).....----------- ( -18.32, z-score = -1.79, R) >droVir3.scaffold_12723 667705 114 + 5802038 UUAAACAAAUACAUUUGAAUAAUGGCAAGCAAUCAGCAAACAAAACAGAAGACUGAUUGCUGGCAAUAAAAAUGGA-CCAUAAAAAGCGAUAACAUCAUAUUUAUAAGCGGCCAG----- .....((((....)))).....((((.((((((((((.............).))))))))).((.(((((.((((.-..................)))).)))))..)).)))).----- ( -19.43, z-score = -1.85, R) >droMoj3.scaffold_6500 16565147 119 - 32352404 UUAAACAAAUACAUUUGAAUAAUGGCUGGCAAUCAGCAAACAAAACAGGAGACUGAUUGCUGGCAAUAAAAAUGGA-CCAUAAAAAGCGAUAACAUCAUAUUUAUAAGCGAAAAGAGACC ...........(((((........((..((((((((....(......)....))))))))..)).....)))))..-.........((.((((........))))..))........... ( -21.32, z-score = -2.49, R) >droGri2.scaffold_15126 240686 114 + 8399593 UUAAACAAAUACAUUUGAAUAAUGGCAAGCAAUGAGCAAACAAAACAGAAGACUGAUUGCUGGCAAUAAAAAUGGA-CCAUAAAAAGCGAUAACAUCAUAUUUAUACGCGGCAAA----- ...........(((((........((.((((((.(((.............).)).)))))).)).....)))))..-.........(((((((........)))).)))......----- ( -16.94, z-score = -0.98, R) >consensus UUAAACAAAUACAUUUGAAUAAUGGCAAGCAAUCAGCAAACAAAACGCAAGACUGAUUGCCGGCAAUAAAAGUGGA_CCAUAAAAAGCGAUAACAUCAUAUUUAUAAGC___________ ...............((((((((((((.(((((((((.............).)))))))).))).......(((....)))............)))..))))))................ (-17.10 = -17.06 + -0.03)
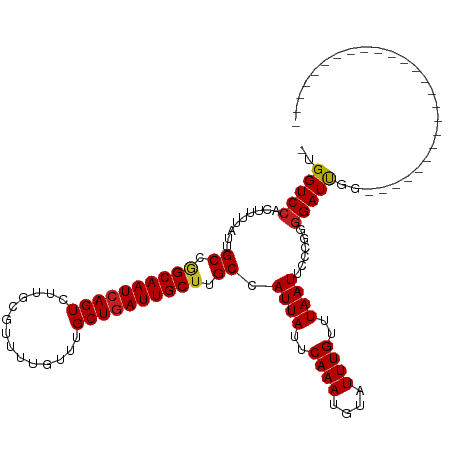
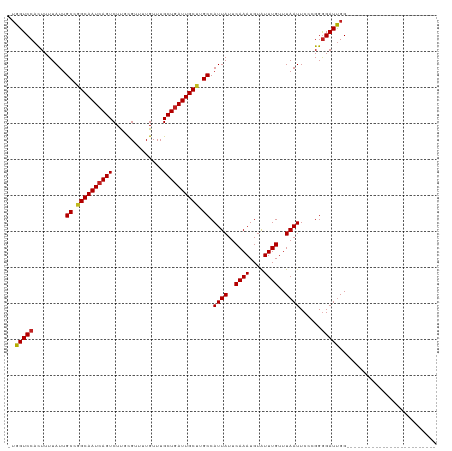
| Location | 13,693,123 – 13,693,216 |
|---|---|
| Length | 93 |
| Sequences | 12 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 87.52 |
| Shannon entropy | 0.23519 |
| G+C content | 0.40369 |
| Mean single sequence MFE | -26.43 |
| Consensus MFE | -21.06 |
| Energy contribution | -21.07 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.908684 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
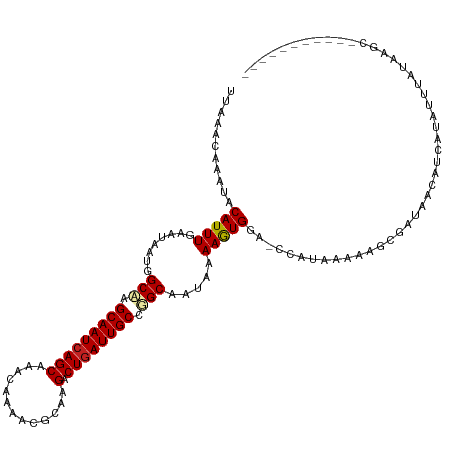
>dm3.chr2L 13693123 93 + 23011544 -UGGUCCACUUUUAUUGCAGGCAAUCAGUCUUGCGUUUUGUUUGCUGAUUGCUUGCCAUUAUUCAAAUGUAUUUGUUUAAUUCCCGGGGAUUGG------------------------- -.(((((.........(((((((((((((...((.....))..))))))))))))).((((..((((....))))..))))......)))))..------------------------- ( -27.50, z-score = -2.65, R) >droSim1.chr2L 13451286 93 + 22036055 -UGGUCCACUUUUAUUGCAGGCAAUCAGUCUUGCGUUUUGUUUGCUGAUUGCUUGCCAUUAUUCAAAUGUAUUUGUUUAAUUCCCGGGGAUUGG------------------------- -.(((((.........(((((((((((((...((.....))..))))))))))))).((((..((((....))))..))))......)))))..------------------------- ( -27.50, z-score = -2.65, R) >droSec1.super_16 1819527 93 + 1878335 -UGGUCCACUUUUAUUGCAGGCAAUCAGUCUUGCGUUUUGUUUGCUGAUUGCUUGCCAUUAUUCAAAUGUAUUUGUUUAAUUCCCGGGGAUUGG------------------------- -.(((((.........(((((((((((((...((.....))..))))))))))))).((((..((((....))))..))))......)))))..------------------------- ( -27.50, z-score = -2.65, R) >droYak2.chr2L 10102597 93 + 22324452 -UGGUCCACUUUUAUUGCAGGCAAUCAGUCUUGCGUUUUGUUUGCUGAUUGCUUGCCAUUAUUCAAAUGUAUUUGUUUAAUUCCCGGGGAUUGG------------------------- -.(((((.........(((((((((((((...((.....))..))))))))))))).((((..((((....))))..))))......)))))..------------------------- ( -27.50, z-score = -2.65, R) >droEre2.scaffold_4929 14866640 93 - 26641161 -UGGUCCACUUUUAUUGCAGGCAAUCAGUCUUGCGUUUUGUUUGCUGAUUGCUUGCCAUUAUUCAAAUGUAUUUGUUUAAUUCCCGGGGAUUGG------------------------- -.(((((.........(((((((((((((...((.....))..))))))))))))).((((..((((....))))..))))......)))))..------------------------- ( -27.50, z-score = -2.65, R) >droAna3.scaffold_12943 1746960 93 + 5039921 -UGGUCCACUUUUAUUGCCGGCAAUCAGUCUUGCGUUUUGUUUGCUGAUUGCUUGCCAUUAUUCAAAUGUGUUUGUUUAAUUCCCGGGGAUUGG------------------------- -.(((((.........((.((((((((((...((.....))..)))))))))).)).((((..((((....))))..))))......)))))..------------------------- ( -24.00, z-score = -1.36, R) >dp4.chr4_group1 3486959 93 - 5278887 -UGGUCCAUUUUUAUUGCCGGCAAUCAGUCUUGCGUUUUGUUUGCUGAUUGCUAGCCAUUAUUCAAAUGUAUUUGUUUAAUUCCCGGGGAUUGG------------------------- -.(((((.........((..(((((((((...((.....))..)))))))))..)).((((..((((....))))..))))......)))))..------------------------- ( -24.20, z-score = -1.56, R) >droPer1.super_5 3414833 93 + 6813705 -UGGUCCAUUUUUAUUGCCGGCAAUCAGUCUUGCGUUUUGUUUGCUGAUUGCUAGCCAUUAUUCAAAUGUAUUUGUUUAAUUCCCGGGGAUUGG------------------------- -.(((((.........((..(((((((((...((.....))..)))))))))..)).((((..((((....))))..))))......)))))..------------------------- ( -24.20, z-score = -1.56, R) >droWil1.scaffold_180772 627998 94 + 8906247 UGGGUCCACUUUUAUUGCCAGCAAUCAGUCUUCUGUUUUGUUUGCUGAUUGCUUGCCAUUAUUCAAAUGUAUUUGUUUAAUUCUUGGGGAUUGG------------------------- ..(((((.........((.((((((((((...(......)...)))))))))).)).((((..((((....))))..))))......)))))..------------------------- ( -23.50, z-score = -1.83, R) >droVir3.scaffold_12723 667740 118 - 5802038 -UGGUCCAUUUUUAUUGCCAGCAAUCAGUCUUCUGUUUUGUUUGCUGAUUGCUUGCCAUUAUUCAAAUGUAUUUGUUUAAUUCUCGGGGAUUGGUUCCGCUGCCAAUGCCGCUGCUCCG -.((..((........((.((((((((((...(......)...)))))))))).)).((((..((((....))))..)))).....((.((((((......)))))).))..))..)). ( -31.60, z-score = -2.11, R) >droMoj3.scaffold_6500 16565187 100 + 32352404 -UGGUCCAUUUUUAUUGCCAGCAAUCAGUCUCCUGUUUUGUUUGCUGAUUGCCAGCCAUUAUUCAAAUGUAUUUGUUUAAUUCCCGGAGAUCGGCGCUGGG------------------ -................(((((...(((....)))........((((((..((.(..((((..((((....))))..))))..).))..))))))))))).------------------ ( -25.40, z-score = -1.15, R) >droGri2.scaffold_15126 240721 111 - 8399593 -UGGUCCAUUUUUAUUGCCAGCAAUCAGUCUUCUGUUUUGUUUGCUCAUUGCUUGCCAUUAUUCAAAUGUAUUUGUUUAAUUCUAGGGGAUUGGU--CACUG-----ACCGCUGGUCCG -...............((((((....(((((((((....((..((.....))..)).((((..((((....))))..))))..)))))))))(((--(...)-----)))))))))... ( -26.80, z-score = -1.49, R) >consensus _UGGUCCACUUUUAUUGCCGGCAAUCAGUCUUGCGUUUUGUUUGCUGAUUGCUUGCCAUUAUUCAAAUGUAUUUGUUUAAUUCCCGGGGAUUGG_________________________ ..(((((.........((.((((((((((..............)))))))))).)).((((..((((....))))..))))......)))))........................... (-21.06 = -21.07 + 0.01)

| Location | 13,693,123 – 13,693,216 |
|---|---|
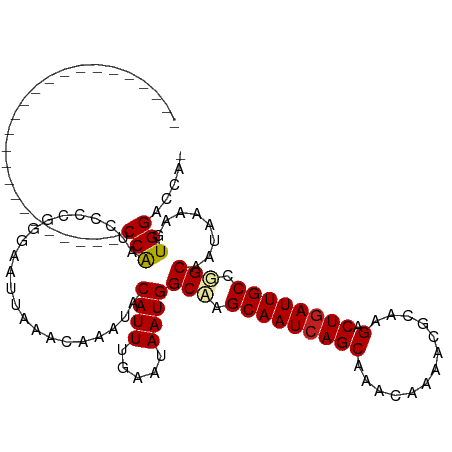
| Length | 93 |
| Sequences | 12 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 87.52 |
| Shannon entropy | 0.23519 |
| G+C content | 0.40369 |
| Mean single sequence MFE | -20.12 |
| Consensus MFE | -16.90 |
| Energy contribution | -17.03 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.742397 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 13693123 93 - 23011544 -------------------------CCAAUCCCCGGGAAUUAAACAAAUACAUUUGAAUAAUGGCAAGCAAUCAGCAAACAAAACGCAAGACUGAUUGCCUGCAAUAAAAGUGGACCA- -------------------------....(((.(................((((.....))))(((.(((((((((.............).)))))))).))).......).)))...- ( -18.42, z-score = -1.35, R) >droSim1.chr2L 13451286 93 - 22036055 -------------------------CCAAUCCCCGGGAAUUAAACAAAUACAUUUGAAUAAUGGCAAGCAAUCAGCAAACAAAACGCAAGACUGAUUGCCUGCAAUAAAAGUGGACCA- -------------------------....(((.(................((((.....))))(((.(((((((((.............).)))))))).))).......).)))...- ( -18.42, z-score = -1.35, R) >droSec1.super_16 1819527 93 - 1878335 -------------------------CCAAUCCCCGGGAAUUAAACAAAUACAUUUGAAUAAUGGCAAGCAAUCAGCAAACAAAACGCAAGACUGAUUGCCUGCAAUAAAAGUGGACCA- -------------------------....(((.(................((((.....))))(((.(((((((((.............).)))))))).))).......).)))...- ( -18.42, z-score = -1.35, R) >droYak2.chr2L 10102597 93 - 22324452 -------------------------CCAAUCCCCGGGAAUUAAACAAAUACAUUUGAAUAAUGGCAAGCAAUCAGCAAACAAAACGCAAGACUGAUUGCCUGCAAUAAAAGUGGACCA- -------------------------....(((.(................((((.....))))(((.(((((((((.............).)))))))).))).......).)))...- ( -18.42, z-score = -1.35, R) >droEre2.scaffold_4929 14866640 93 + 26641161 -------------------------CCAAUCCCCGGGAAUUAAACAAAUACAUUUGAAUAAUGGCAAGCAAUCAGCAAACAAAACGCAAGACUGAUUGCCUGCAAUAAAAGUGGACCA- -------------------------....(((.(................((((.....))))(((.(((((((((.............).)))))))).))).......).)))...- ( -18.42, z-score = -1.35, R) >droAna3.scaffold_12943 1746960 93 - 5039921 -------------------------CCAAUCCCCGGGAAUUAAACAAACACAUUUGAAUAAUGGCAAGCAAUCAGCAAACAAAACGCAAGACUGAUUGCCGGCAAUAAAAGUGGACCA- -------------------------....(((.(................((((.....))))((..(((((((((.............).))))))))..)).......).)))...- ( -17.22, z-score = -0.66, R) >dp4.chr4_group1 3486959 93 + 5278887 -------------------------CCAAUCCCCGGGAAUUAAACAAAUACAUUUGAAUAAUGGCUAGCAAUCAGCAAACAAAACGCAAGACUGAUUGCCGGCAAUAAAAAUGGACCA- -------------------------(((.(((...)))............((((.....))))((..(((((((((.............).))))))))..))........)))....- ( -18.82, z-score = -1.55, R) >droPer1.super_5 3414833 93 - 6813705 -------------------------CCAAUCCCCGGGAAUUAAACAAAUACAUUUGAAUAAUGGCUAGCAAUCAGCAAACAAAACGCAAGACUGAUUGCCGGCAAUAAAAAUGGACCA- -------------------------(((.(((...)))............((((.....))))((..(((((((((.............).))))))))..))........)))....- ( -18.82, z-score = -1.55, R) >droWil1.scaffold_180772 627998 94 - 8906247 -------------------------CCAAUCCCCAAGAAUUAAACAAAUACAUUUGAAUAAUGGCAAGCAAUCAGCAAACAAAACAGAAGACUGAUUGCUGGCAAUAAAAGUGGACCCA -------------------------....(((.(................((((.....))))((.((((((((((.............).))))))))).)).......).))).... ( -18.42, z-score = -2.17, R) >droVir3.scaffold_12723 667740 118 + 5802038 CGGAGCAGCGGCAUUGGCAGCGGAACCAAUCCCCGAGAAUUAAACAAAUACAUUUGAAUAAUGGCAAGCAAUCAGCAAACAAAACAGAAGACUGAUUGCUGGCAAUAAAAAUGGACCA- (((....((.((....)).))(((.....))))))...............(((((........((.((((((((((.............).))))))))).)).....))))).....- ( -26.04, z-score = -1.12, R) >droMoj3.scaffold_6500 16565187 100 - 32352404 ------------------CCCAGCGCCGAUCUCCGGGAAUUAAACAAAUACAUUUGAAUAAUGGCUGGCAAUCAGCAAACAAAACAGGAGACUGAUUGCUGGCAAUAAAAAUGGACCA- ------------------.(((((((.(((..((((..((((..((((....))))..))))..))))..))).))........(((....)))...)))))................- ( -27.20, z-score = -2.64, R) >droGri2.scaffold_15126 240721 111 + 8399593 CGGACCAGCGGU-----CAGUG--ACCAAUCCCCUAGAAUUAAACAAAUACAUUUGAAUAAUGGCAAGCAAUGAGCAAACAAAACAGAAGACUGAUUGCUGGCAAUAAAAAUGGACCA- ..(.((((((((-----(((((--.(((.((.....))......((((....)))).....))))..((.....))..............)))))))))))))...............- ( -22.80, z-score = -1.43, R) >consensus _________________________CCAAUCCCCGGGAAUUAAACAAAUACAUUUGAAUAAUGGCAAGCAAUCAGCAAACAAAACGCAAGACUGAUUGCCGGCAAUAAAAGUGGACCA_ .........................(((......................((((.....))))(((.(((((((((.............).)))))))).)))........)))..... (-16.90 = -17.03 + 0.13)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:38:32 2011