| Sequence ID | dm3.chr2L |
|---|---|
| Location | 13,673,380 – 13,673,510 |
| Length | 130 |
| Max. P | 0.946070 |

| Location | 13,673,380 – 13,673,510 |
|---|---|
| Length | 130 |
| Sequences | 12 |
| Columns | 146 |
| Reading direction | reverse |
| Mean pairwise identity | 75.26 |
| Shannon entropy | 0.53543 |
| G+C content | 0.46190 |
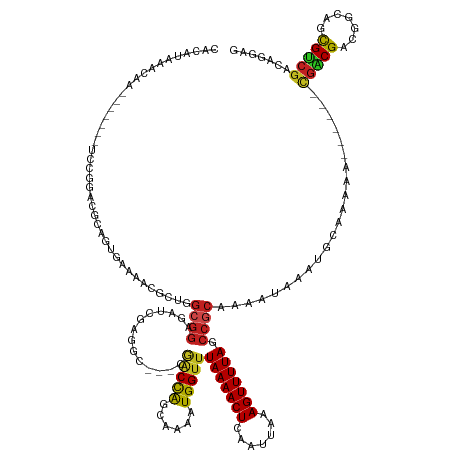
| Mean single sequence MFE | -35.06 |
| Consensus MFE | -16.94 |
| Energy contribution | -16.65 |
| Covariance contribution | -0.29 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.52 |
| SVM RNA-class probability | 0.946070 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
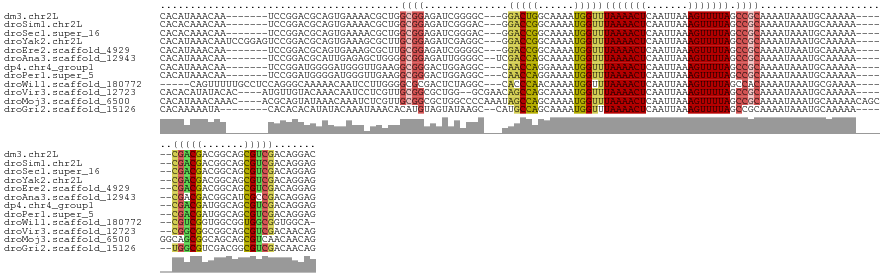
>dm3.chr2L 13673380 130 - 23011544 CACAUAAACAA-------UCCGGACGCAGUGAAAACGCUGGCGGAGAUCGGGGC---GGACUGGCAAAAUGGUUUAAAACUCAAUUAAAGUUUUAGCCGCAAAAUAAAUGCAAAAA------CGACGACGGCAGCGUCGACAGGAC ...........-------(((.(((...((....))((((.((..(.(((.(((---.(((((......)))))(((((((.......))))))))))(((.......))).....------))))..)).)))))))....))). ( -34.90, z-score = -2.00, R) >droSim1.chr2L 13433077 130 - 22036055 CACACAAACAA-------UCCGGACGCAGUGAAAACGCUGGCGGAGAUCGGGAC---GGACCGGCAAAAUGGUUUAAAACUCAAUUAAAGUUUUAGCCGCAAAAUAAAUGCAAAAA------CGACGACGGCAGCGUCGACAGGAG ...........-------(((.(((((.((....))((((.((..(.((((...---...))))......((((.((((((.......))))))))))(((.......))).....------)..)).)))).)))))....))). ( -35.70, z-score = -2.26, R) >droSec1.super_16 1799837 130 - 1878335 CACACAAACAA-------UCCGGACGCAGUGAAAACGCUGGCGGAGAUCGGGAC---GGACCGGCAAAAUGGUUUAAAACUCAAUUAAAGUUUUAGCCGCAAAAUAAAUGCAAAAA------CGACGACGGCAGCGUCGACAGGAG ...........-------(((.(((((.((....))((((.((..(.((((...---...))))......((((.((((((.......))))))))))(((.......))).....------)..)).)))).)))))....))). ( -35.70, z-score = -2.26, R) >droYak2.chr2L 10082878 137 - 22324452 CACAUAAACAAUCCGGAGUCCGGACGCAGUGAAAGCGCUUGCGGAGAUCGAGGC---GGACCGGCAAAAUGGUUUAAAACUCAAUUAAAGUUUUAGCCGCAAAAUAAAUGCAAAAA------CGACGACGGCAGCGUCGACAGGAG ...........(((...(((..(((((.((....))(((..((..(.....(((---.(((((......)))))(((((((.......))))))))))(((.......))).....------)..))..))).)))))))).))). ( -37.30, z-score = -1.22, R) >droEre2.scaffold_4929 14846210 130 + 26641161 CACAUAAACAA-------UCCGGACGCAGUGAAAGCGCUUGCGGAGAUCGGGGC---GGACCGGCAAAAUGGUUUAAAACUCAAUUAAAGUUUUAGCCGCAAAAUAAAUGCAAAAA------CGACGACGGCAGCGUCGACAGGAG ...........-------(((.(((((.((....))(((..((..(.....(((---.(((((......)))))(((((((.......))))))))))(((.......))).....------)..))..))).)))))....))). ( -34.70, z-score = -1.30, R) >droAna3.scaffold_12943 1724688 131 - 5039921 CACAUAAACAA-------UCCGGACGCAUUGAGAGCUGGGGCGGAGAUUGGGGC--UCGACCAGCAAAAUGGUUUAAAACUCAAUUAAAGUUUUAGCCGCAAAAUAAAUGCAAAAA------CGACGACGGCAUCGCCGACAGGAG ...........-------(((....(((((..(.((((((((...(((((((..--..(((((......))))).....)))))))...)))))))))........))))).....------......((((...))))...))). ( -33.60, z-score = -1.54, R) >dp4.chr4_group1 3460701 130 + 5278887 CACAUAAACAA-------UCCGGAUGGGGAUGGGUUGAAGGCGGGACUGGAGGC---CAACCAGGAAAAUGGUUUAAAACUCAAUUAAAGUUUUAGCCGCAAAAUAAAUGCAAAAA------CGACGAUGGCAGCGUCGACAGGAG ........(((-------((((........)))))))..(((((((((.(((..---.(((((......))))).....)))......)))))).)))(((.......))).....------(((((.......)))))....... ( -31.20, z-score = -1.23, R) >droPer1.super_5 3388121 130 - 6813705 CACAUAAACAA-------UCCGGAUGGGGAUGGGUUGAAGGCGGGACUGGAGGC---CAACCAGGAAAAUGGUUUAAAACUCAAUUAAAGUUUUAGCCGCAAAAUAAAUGCAAAAA------CGACGAUGGCAGCGUCGACAGGAG ........(((-------((((........)))))))..(((((((((.(((..---.(((((......))))).....)))......)))))).)))(((.......))).....------(((((.......)))))....... ( -31.20, z-score = -1.23, R) >droWil1.scaffold_180772 712346 131 - 8906247 -----CAGUUUUUGCCUCCAGGGCAAAAACAAUCCUUGGGGCGCGACUCUAGGC---CACCCAACAAAAUGGUUUAAAACUCAAUUAAAGUUUUAGCCACAAAAUAAAUGCGAAAA------CGUCGGUGGCGGUGGCGGUGGCA- -----..(((((((((.....))))))))).....((((((.....))))))((---((((.............(((((((.......)))))))(((((....((...(((....------)))...))...))))))))))).- ( -42.30, z-score = -1.76, R) >droVir3.scaffold_12723 637538 134 + 5802038 CACACAUAUACAC----AUGUUGUACAAACAAUCCUCGUUGCGGCGCUGG--GCGAACAGCCAGCAAAAUGGUUUAAAACUCAAUUAAAGUUUUAGCCGCAAAAUAAAUGCAAAAA------CGGCGGCGGCAGCGUCGACAACAG .............----.((((((............((((((.(((((((--((....(((((......)))))(((((((.......))))))))))(((.......))).....------)))).)).))))))...)))))). ( -38.56, z-score = -2.35, R) >droMoj3.scaffold_6500 16528350 142 - 32352404 CACAUAAACAAAC----ACGCAGUAUAAACAAAUCUCGUUGCGGCGCUGGCCCCAAAUAGCCAGCAAAAUGGUUUAAAACUCAAUUAAAGUUUUAGCCGCAAAAUAAAUGCAAAAACAGCGGCAGCGGCAGCAGCGUCAACAACAG .............----((((.((....)).......(((((.((((((((........)))))).........(((((((.......)))))))(((((..................))))).)).))))).))))......... ( -40.07, z-score = -3.65, R) >droGri2.scaffold_15126 212586 130 + 8399593 CACAAAAAUA--------CACACACAUAUACAAAUAAACACAUGUAGUAUAAGC--CAUGCCAGCAAAAUGGUUUAAAACUCAAUUAAAGUUUUAGCCGCAAAAUAAAUGCAAAAA------UGGCGUCGACGGCGUCGACAACAG ..........--------........(((((...............))))).((--(((...........((((.((((((.......))))))))))(((.......)))....)------))))((((((...))))))..... ( -25.46, z-score = -2.36, R) >consensus CACAUAAACAA_______UCCGGACGCAGUGAAAACGCUGGCGGAGAUCGAGGC___CGACCAGCAAAAUGGUUUAAAACUCAAUUAAAGUUUUAGCCGCAAAAUAAAUGCAAAAA______CGACGACGGCAGCGUCGACAGGAG ........................................((((..............(((((......)))))(((((((.......))))))).))))......................(((((.......)))))....... (-16.94 = -16.65 + -0.29)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:38:29 2011