| Sequence ID | dm3.chr2L |
|---|---|
| Location | 13,644,903 – 13,644,994 |
| Length | 91 |
| Max. P | 0.878822 |

| Location | 13,644,903 – 13,644,994 |
|---|---|
| Length | 91 |
| Sequences | 10 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 78.32 |
| Shannon entropy | 0.45942 |
| G+C content | 0.42436 |
| Mean single sequence MFE | -15.93 |
| Consensus MFE | -9.88 |
| Energy contribution | -9.95 |
| Covariance contribution | 0.07 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.878822 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
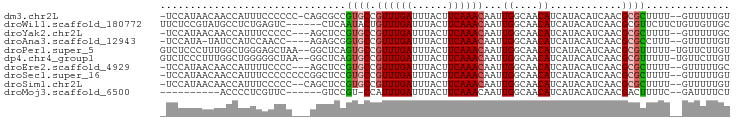
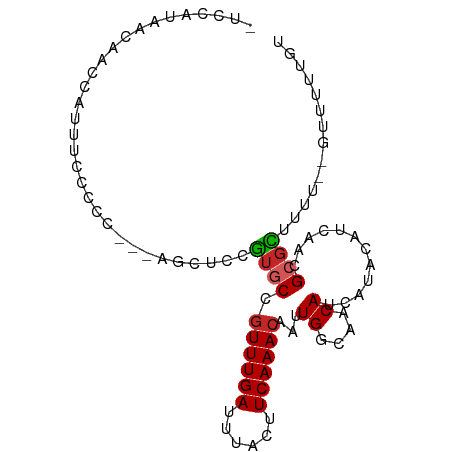
>dm3.chr2L 13644903 91 + 23011544 -UCCAUAACAACCAUUUCCCCCC-CAGCGCCGUGCCGUUUGAUUUACUUCAAACAAUUGGCAACAUCAUACAUCAACGCGCUUUU--GUUUUUGU -......................-.(((((.((...((((((......))))))...((....))..........)))))))...--........ ( -15.80, z-score = -1.98, R) >droWil1.scaffold_180772 8107785 89 - 8906247 UUCUCCGUAUGCCUCUGAGUC------CUCAAUACUGUUUGAUUUACUUCAAACAAUUGGCAACAUCAUACAUCAACGCGUUCUUCUGUUGUUGC ......((.((((..(((...------.)))....(((((((......)))))))...))))))......((.(((((........))))).)). ( -17.10, z-score = -2.09, R) >droYak2.chr2L 10063617 89 + 22324452 -UCCAUAACAACCAUUUCCCCC---AGCUCCGUGCCGUUUGAUUUACUUCAAACAAUUGGCAACAUCAUACAUCAACGCGCUUUU--GUUUUUGC -.....................---......((((.((((((......))))))...((....))............))))....--........ ( -13.20, z-score = -1.40, R) >droAna3.scaffold_12943 1703281 87 + 5039921 -UCCAUA-UAUCCAUCCAACC----AGAGCGGUGCCGUUUGAUUUACUUCAAACAAUUGGCAACAUCAUACAUCAACGCGCCUUU--GUUUUUGU -......-..........((.----((((((((((.((((((......))))))...((....))............)))))...--))))).)) ( -17.70, z-score = -1.95, R) >droPer1.super_5 3366675 92 + 6813705 GUCUCCCUUUGGCUGGGAGCUAA--GGCUCAGUGCCGUUUGAUUUACUUCAAACAAUUGGCAACAUCAUACAUCAACGCGUUUUU-UGUUCUUGU ...((((.......))))(((((--(((.....)))((((((......))))))..)))))........(((..((((.......-))))..))) ( -23.10, z-score = -1.48, R) >dp4.chr4_group1 3439445 92 - 5278887 GUCUCCCUUUGGCUGGGGGCUAA--GGCUCAGUGCCGUUUGAUUUACUUCAAACAAUUGGCAACAUCAUACAUCAACGCGUUUUU-UGUUCUUGU ....((((......))))(((((--(((.....)))((((((......))))))..)))))........(((..((((.......-))))..))) ( -22.90, z-score = -1.17, R) >droEre2.scaffold_4929 14826965 89 - 26641161 -UCCAUAACAACCAUUUUCCCC---AGCUCCGUGCCGUUUGAUUUACUUCAAACAAUUGGCAACAUCAUACAUCAACGCGCUUUU--GUUUUUGC -.....................---......((((.((((((......))))))...((....))............))))....--........ ( -13.20, z-score = -1.35, R) >droSec1.super_16 1780493 92 + 1878335 -UCCAUAACAACCAUUUCCCCCCCCGGCUCCGUGCCGUUUGAUUUACUUCAAACAAUUGGCAACAUCAUACAUCAACGCGCUUUU--GUUUUUGU -..........(((...........(((.....)))((((((......))))))...))).........(((..((((......)--)))..))) ( -14.90, z-score = -1.39, R) >droSim1.chr2L 13413412 90 + 22036055 -UCCAUAACAACCAUUUCCCCC--CAGCUCCGUGCCGUUUGAUUUACUUCAAACAAUUGGCAACAUCAUACAUCAACGCGCUUUU--GUUUUUGU -.....................--.......((((.((((((......))))))...((....))............))))....--........ ( -13.20, z-score = -1.54, R) >droMoj3.scaffold_6500 16499769 76 + 32352404 ----------ACCCCUCGUUC------GUCCGU-CCAUUUGAUUUACUUCAAACAAUUGGCAACAUCAUACAUCAACGACUUUUC--GAUUUUCU ----------.........((------(...((-(..(((((......)))))....((....))............)))....)--))...... ( -8.20, z-score = -0.83, R) >consensus _UCCAUAACAACCAUUUCCCCC___AGCUCCGUGCCGUUUGAUUUACUUCAAACAAUUGGCAACAUCAUACAUCAACGCGCUUUU__GUUUUUGU ...............................((((.((((((......))))))...((....))............)))).............. ( -9.88 = -9.95 + 0.07)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:38:28 2011