| Sequence ID | dm3.chr2L |
|---|---|
| Location | 13,632,865 – 13,632,963 |
| Length | 98 |
| Max. P | 0.917987 |

| Location | 13,632,865 – 13,632,963 |
|---|---|
| Length | 98 |
| Sequences | 9 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 75.88 |
| Shannon entropy | 0.50191 |
| G+C content | 0.40567 |
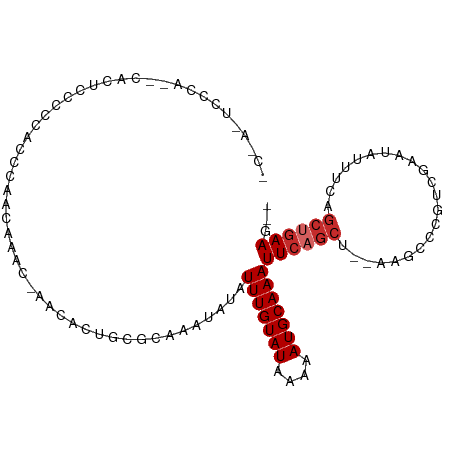
| Mean single sequence MFE | -11.62 |
| Consensus MFE | -7.39 |
| Energy contribution | -7.95 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.917987 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
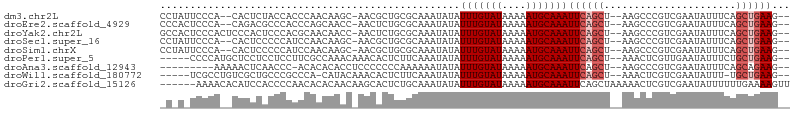
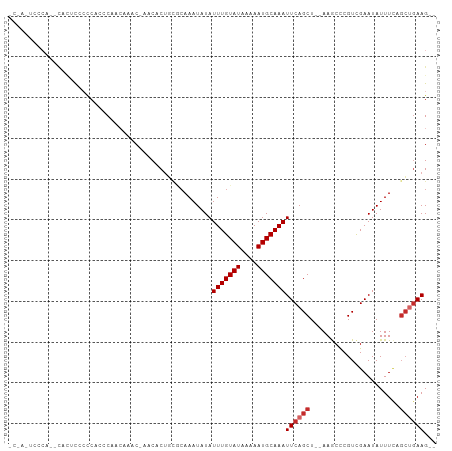
>dm3.chr2L 13632865 98 - 23011544 CCUAUUCCCA--CACUCUACCACCCAACAAGC-AACGCUGCGCAAAUAUAUUUGUAUAAAAAUGCAAAUUCAGCU--AAGCCCGUCGAAUAUUUCAGCUGAAG-- ..........--..................((-(....))).........(((((((....)))))))(((((((--..................))))))).-- ( -12.87, z-score = -1.01, R) >droEre2.scaffold_4929 14814779 98 + 26641161 CCCACUCCCA--CAGACGCCCACCCAGCAACC-AACUCUGCGCAAAUAUAUUUGUAUAAAAAUGCAAAUUCAGCU--AAGCCCGUCGAAUAUUUCAGCUGAAG-- ..........--..((((..(.....(((...-.....)))((......((((((((....))))))))...)).--..)..)))).................-- ( -13.00, z-score = -1.16, R) >droYak2.chr2L 10051678 100 - 22324452 GCCACUCCCACUCCCACUCCCACGCAACAACC-AACUCUGCGCAAAUAUAUUUGUAUAAAAAUGCAAAUUCAGCU--AAGCCCGUCGAAUAUUUCAGCUGAAG-- ......................((((......-.....))))........(((((((....)))))))(((((((--..................))))))).-- ( -13.27, z-score = -1.83, R) >droSec1.super_16 1768495 98 - 1878335 CCUAUUCCCA--CACUCCCCCAUCCAACAAGC-AACGCUGCGCAAAUAUAUUUGUAUAAAAAUGCAAAUUCAGCU--AAGCCCGUCGAAUAUUUCAGCUGAAG-- ..........--..................((-(....))).........(((((((....)))))))(((((((--..................))))))).-- ( -12.87, z-score = -1.32, R) >droSim1.chrX 177089 98 + 17042790 CCUAUUCCCA--CACUCCCCCAUCCAACAAGC-AACGCUGCGCAAAUAUAUUUGUAUAAAAAUGCAAAUUCAGCU--AAGCCCGUCGAAUAUUUCAGCUGAAG-- ..........--..................((-(....))).........(((((((....)))))))(((((((--..................))))))).-- ( -12.87, z-score = -1.32, R) >droPer1.super_5 3353045 96 - 6813705 -----CCCCAUGCUCCUCCUCCUUCGCCAAACAAACACUCUUCAAAUAUAUUUGUAUAAAAAUGCAAAUUCAGCU--AAACUCGUUGAAUAUUUCUGCUGAAG-- -----................(((((.((......................((((((....))))))(((((((.--......))))))).....)).)))))-- ( -12.50, z-score = -2.94, R) >droAna3.scaffold_12943 1691188 91 - 5039921 ---------AAAAACUCAACCC-ACACACACCUCCCCCCCAAAAAAUAUAUUUGUAUAAAAAUGCAAAUUCAGCU--AAGCCCGUCGAAUAUUUCAGCAGAAG-- ---------.............-...........................(((((((....)))))))(((.(((--..................))).))).-- ( -5.27, z-score = -1.03, R) >droWil1.scaffold_180772 8077810 94 + 8906247 -----UCGCCUGUCGCUGCCCGCCCA-CAUACAAACACUCUUCAAAUAUAUUUGUAUAAAAAUGCAAAUUCAGCU--AAACUCGUCGAAUAUUU-UGCUGAAG-- -----..(..(((.((.....))..)-))..)..................(((((((....)))))))((((((.--.................-.)))))).-- ( -10.71, z-score = -0.11, R) >droGri2.scaffold_15126 163840 99 + 8399593 ------AAAACACAUCCACCCCAACACACAACAAGCACUCUGCAAAUAUAUUUGUAUAAAAAUGCAAAUUCAGCUAAAAACUCGUCGAAUAUUUUUUGAAAAGUU ------............................((.....))......((((((((....)))))))).........((((..(((((.....)))))..)))) ( -11.20, z-score = -1.54, R) >consensus _C_A_UCCCA__CACUCCCCCACCCAACAAAC_AACACUGCGCAAAUAUAUUUGUAUAAAAAUGCAAAUUCAGCU__AAGCCCGUCGAAUAUUUCAGCUGAAG__ ..................................................(((((((....)))))))((((((......................))))))... ( -7.39 = -7.95 + 0.56)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:38:27 2011