| Sequence ID | dm3.chr2L |
|---|---|
| Location | 13,570,208 – 13,570,301 |
| Length | 93 |
| Max. P | 0.984214 |

| Location | 13,570,208 – 13,570,301 |
|---|---|
| Length | 93 |
| Sequences | 10 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 73.89 |
| Shannon entropy | 0.51672 |
| G+C content | 0.37118 |
| Mean single sequence MFE | -15.86 |
| Consensus MFE | -6.66 |
| Energy contribution | -6.96 |
| Covariance contribution | 0.30 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.16 |
| SVM RNA-class probability | 0.984214 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
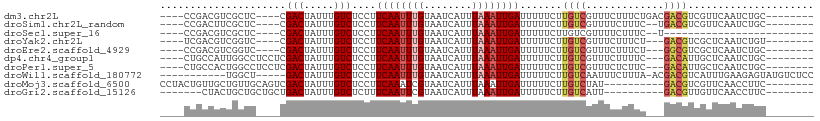
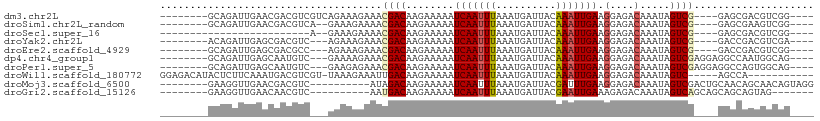
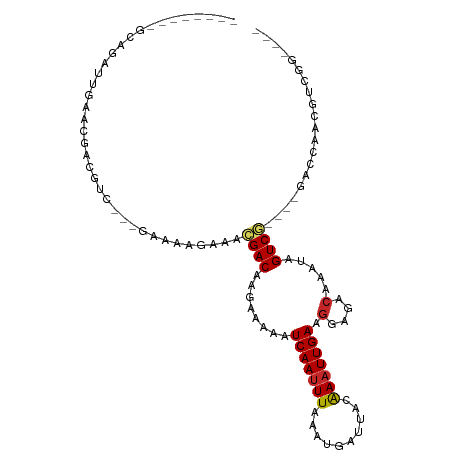
>dm3.chr2L 13570208 93 + 23011544 ----CCGACGUCGCUC----CGACUAUUUGUCUCCUUCAAUUUGUAAUCAUUUAAAUUGAUUUUUCUUGUCGUUUCUUUCUGACGACGUCGUUCAAUCUGC-------- ----.((((((((.((----.(((.....)))....((((((((........)))))))).....................))))))))))..........-------- ( -21.90, z-score = -3.94, R) >droSim1.chr2L_random 516206 91 + 909653 ----CCGACUUCGCUC----CGACUAUUUGUCUCCUUCAAUUUGUAAUCAUUUAAAUUGAUUUUUCUUGUCGUUUUCUUUC--UGACGUCGUUCAAUCUGC-------- ----.((((.((....----.(((.....)))....((((((((........)))))))).....................--.)).))))..........-------- ( -13.90, z-score = -2.20, R) >droSec1.super_16 1707546 74 + 1878335 ----CCGACGUCGCUC----CGACUAUUUGUCUCCUUCAAUUUGUAAUCAUUUAAAUUGAUUUUUCUUGUCGUUUUCUUUC--U------------------------- ----.((((((((...----))))............((((((((........))))))))........)))).........--.------------------------- ( -12.90, z-score = -3.86, R) >droYak2.chr2L 9989415 90 + 22324452 ----UCGACGUCGGUC----CGACUAUUUGUCUCCUUCAAUUUGUAAUCAUUUAAAUUGAUUUUUCUUGUCGUUUCUUUCU---GACGUCGCUCAAUCUGU-------- ----.(((((((((..----.(((.....)))....((((((((........))))))))...................))---)))))))..........-------- ( -21.70, z-score = -4.33, R) >droEre2.scaffold_4929 14754426 90 - 26641161 ----CCGACGUCGGUC----CGACUAUUUGUCUCCUUCAAUUUGUAAUCAUUUAAAUUGAUUUUUCUUGUCGUUUCUUUCU---GGCGUCGCUCAAUCUGC-------- ----.(((((((((..----.(((.....)))....((((((((........))))))))...................))---)))))))..........-------- ( -21.30, z-score = -4.02, R) >dp4.chr4_group1 3336495 94 - 5278887 ----CUGCCAUUGGCCUCCUCGACUAUUUGUCUCCUUCAAUUUGUAAUCAUUUAAAUUGAUUUUUCUUGUCGUUUCUUUUC---GACAUUGCUCAAUCUGC-------- ----..((.((((((......(((.....)))....((((((((........)))))))).......(((((........)---))))..)).))))..))-------- ( -16.10, z-score = -3.05, R) >droPer1.super_5 3263536 94 + 6813705 ----CUGCCACUGGCCUCCUCGACUAUUUGUCUCCUUCAAUUUGUAAUCAUUUAAAUUGAUUUUUCUUGUCGUUUCUCUUC---GACAUUGCUCAAUCUGC-------- ----..(((...)))......(((.....)))....((((((((........)))))))).......(((((........)---)))).............-------- ( -14.70, z-score = -2.55, R) >droWil1.scaffold_180772 7982259 92 - 8906247 -----------UGGCU-----GACUAUUUGUCUCCUUCAAUUUGUAAUCAUUUAAAUUGAUUUUUCUUGUCAAUUUCUUUA-ACGACGUCAUUUGAAGAGUAUGUCUCC -----------.(((.-----(((.....)))..((((((..(((........((((((((.......)))))))).....-...)))....)))))).....)))... ( -15.69, z-score = -1.42, R) >droMoj3.scaffold_6500 16372262 91 + 32352404 CCUACUGUUGCUGUUGCAGUCGACUAUUUGUCUCCUUCAAAUCGUAAUCAUUUAAAUUGAUUUUUCUUGUCUAU----------GACGUCGUUCAACCUUC-------- ...(((((.......)))))((((.....(((.....(((.....(((((.......)))))....))).....----------)))))))..........-------- ( -11.90, z-score = -0.96, R) >droGri2.scaffold_15126 64062 84 - 8399593 -------CUACUGCUGCUGCUGACUAUUUGUCUCUUUCAAUUCGUAAUCAUUUAAAUUGAUUUUUCUUGUCAUU----------GACGUUGUUCAACCUUC-------- -------.....((....))(((.((..((((.....(((.....(((((.......)))))....))).....----------)))).)).)))......-------- ( -8.50, z-score = -0.38, R) >consensus ____CCGACGUCGGUC____CGACUAUUUGUCUCCUUCAAUUUGUAAUCAUUUAAAUUGAUUUUUCUUGUCGUUUCUUUUC___GACGUCGUUCAAUCUGC________ .....................(((.....)))....((((((((........))))))))................................................. ( -6.66 = -6.96 + 0.30)
| Location | 13,570,208 – 13,570,301 |
|---|---|
| Length | 93 |
| Sequences | 10 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 73.89 |
| Shannon entropy | 0.51672 |
| G+C content | 0.37118 |
| Mean single sequence MFE | -16.23 |
| Consensus MFE | -7.72 |
| Energy contribution | -7.82 |
| Covariance contribution | 0.10 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.935396 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
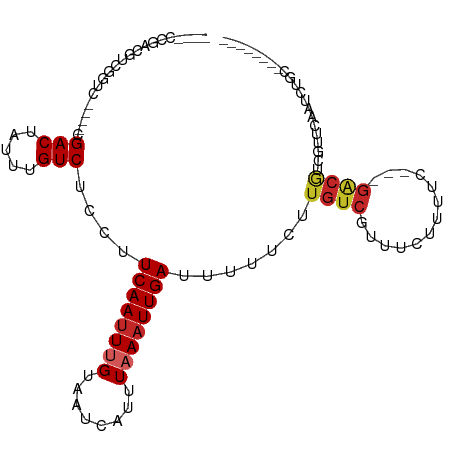
>dm3.chr2L 13570208 93 - 23011544 --------GCAGAUUGAACGACGUCGUCAGAAAGAAACGACAAGAAAAAUCAAUUUAAAUGAUUACAAAUUGAAGGAGACAAAUAGUCG----GAGCGACGUCGG---- --------..........(((((((((..........((((........(((((((..........))))))).(....).....))))----..))))))))).---- ( -24.10, z-score = -3.60, R) >droSim1.chr2L_random 516206 91 - 909653 --------GCAGAUUGAACGACGUCA--GAAAGAAAACGACAAGAAAAAUCAAUUUAAAUGAUUACAAAUUGAAGGAGACAAAUAGUCG----GAGCGAAGUCGG---- --------..........((((.((.--....))...((((........(((((((..........))))))).(....).....))))----.......)))).---- ( -15.40, z-score = -1.64, R) >droSec1.super_16 1707546 74 - 1878335 -------------------------A--GAAAGAAAACGACAAGAAAAAUCAAUUUAAAUGAUUACAAAUUGAAGGAGACAAAUAGUCG----GAGCGACGUCGG---- -------------------------.--.........((((........(((((((..........))))))).(....).....((((----...)))))))).---- ( -13.40, z-score = -2.83, R) >droYak2.chr2L 9989415 90 - 22324452 --------ACAGAUUGAGCGACGUC---AGAAAGAAACGACAAGAAAAAUCAAUUUAAAUGAUUACAAAUUGAAGGAGACAAAUAGUCG----GACCGACGUCGA---- --------..........(((((((---.........((((........(((((((..........))))))).(....).....))))----....))))))).---- ( -19.82, z-score = -2.81, R) >droEre2.scaffold_4929 14754426 90 + 26641161 --------GCAGAUUGAGCGACGCC---AGAAAGAAACGACAAGAAAAAUCAAUUUAAAUGAUUACAAAUUGAAGGAGACAAAUAGUCG----GACCGACGUCGG---- --------..........(((((..---.........((((........(((((((..........))))))).(....).....))))----......))))).---- ( -15.13, z-score = -1.25, R) >dp4.chr4_group1 3336495 94 + 5278887 --------GCAGAUUGAGCAAUGUC---GAAAAGAAACGACAAGAAAAAUCAAUUUAAAUGAUUACAAAUUGAAGGAGACAAAUAGUCGAGGAGGCCAAUGGCAG---- --------((..((((.((..((((---(........))))).......(((((((..........)))))))....(((.....)))......)))))).))..---- ( -17.50, z-score = -2.54, R) >droPer1.super_5 3263536 94 - 6813705 --------GCAGAUUGAGCAAUGUC---GAAGAGAAACGACAAGAAAAAUCAAUUUAAAUGAUUACAAAUUGAAGGAGACAAAUAGUCGAGGAGGCCAGUGGCAG---- --------((..((((.((..((((---(........))))).......(((((((..........)))))))....(((.....)))......)))))).))..---- ( -17.50, z-score = -2.21, R) >droWil1.scaffold_180772 7982259 92 + 8906247 GGAGACAUACUCUUCAAAUGACGUCGU-UAAAGAAAUUGACAAGAAAAAUCAAUUUAAAUGAUUACAAAUUGAAGGAGACAAAUAGUC-----AGCCA----------- (((((.....)))))...((((((.((-((...(((((((.........)))))))...)))).))........(....).....)))-----)....----------- ( -15.80, z-score = -1.59, R) >droMoj3.scaffold_6500 16372262 91 - 32352404 --------GAAGGUUGAACGACGUC----------AUAGACAAGAAAAAUCAAUUUAAAUGAUUACGAUUUGAAGGAGACAAAUAGUCGACUGCAACAGCAACAGUAGG --------....((((..((..(((----------...)))......(((((.......))))).))..(((.((..(((.....)))..)).)))))))......... ( -13.60, z-score = -0.77, R) >droGri2.scaffold_15126 64062 84 + 8399593 --------GAAGGUUGAACAACGUC----------AAUGACAAGAAAAAUCAAUUUAAAUGAUUACGAAUUGAAAGAGACAAAUAGUCAGCAGCAGCAGUAG------- --------....(((((.....(((----------..(..(((....(((((.......))))).....)))..)..)))......)))))...........------- ( -10.00, z-score = -0.35, R) >consensus ________GCAGAUUGAACGACGUC___GAAAAGAAACGACAAGAAAAAUCAAUUUAAAUGAUUACAAAUUGAAGGAGACAAAUAGUCG____GACCAACGUCGG____ .....................................((((........(((((((..........))))))).(....).....)))).................... ( -7.72 = -7.82 + 0.10)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:38:18 2011