
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 1,262,236 – 1,262,326 |
| Length | 90 |
| Max. P | 0.770794 |

| Location | 1,262,236 – 1,262,326 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 77.91 |
| Shannon entropy | 0.38377 |
| G+C content | 0.45061 |
| Mean single sequence MFE | -24.30 |
| Consensus MFE | -16.70 |
| Energy contribution | -18.70 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.770794 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
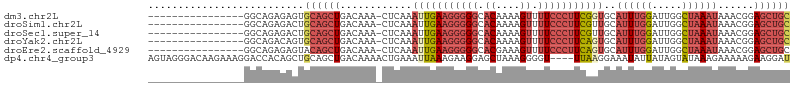
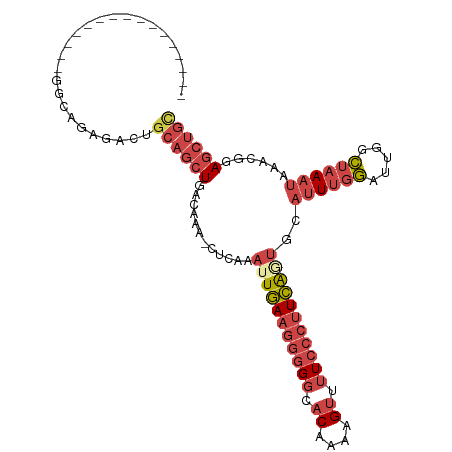
>dm3.chr2L 1262236 90 + 23011544 ----------------GGCAGAGAGUGCAGCUGACAAA-CUCAAAUUGAAGGGGGCACAAAAGUUUUCCCUUCGGUGCAUUUGGAUUGGCUAAAUAAACGGAGCUGC ----------------..........((((((......-.....(((((((((((.((....)).)))))))))))..((((((.....))))))......)))))) ( -25.50, z-score = -0.98, R) >droSim1.chr2L 1234252 90 + 22036055 ----------------GGCAGAGACUGCAGCUGACAAA-CUCAAAUUGAAGGGGGCACAAAAGUUUUCCCUUCGUUGCAUUUGGAUUGGCUAAAUAAACGGAGCUGC ----------------..........((((((......-........((((((((.((....)).))))))))(((..((((((.....)))))).)))..)))))) ( -25.90, z-score = -1.37, R) >droSec1.super_14 1212827 90 + 2068291 ----------------GGCAGAGACUGCAGCUGACAAA-CUCAAAUUGAAGGGGGCACAAAAGUUUUCCCUUCGUUGCAUUUGGAUUGGCUAAAUAAACGGAGCUGC ----------------..........((((((......-........((((((((.((....)).))))))))(((..((((((.....)))))).)))..)))))) ( -25.90, z-score = -1.37, R) >droYak2.chr2L 1237346 90 + 22324452 ----------------GGCAGACAGUGCAGCUGACAAA-CUCAAAUUGAAGGGGGCACAAAAGUUUUCCCUUCAGUGCAUUUGGAUUGGCUAAAUAAACGGAGCUGC ----------------.((((.(((.....))).....-(((..(((((((((((.((....)).)))))))))))..((((((.....)))))).....))))))) ( -26.40, z-score = -1.39, R) >droEre2.scaffold_4929 1302851 90 + 26641161 ----------------GGCAGAGAGUACAGCUGACAAA-CUCAAAUUGAAGGGGGCACGAAAGUUUUCCCUUCAGUGCAUUUGGAUUGGCUAAAUAAACGGAGCUGC ----------------.((((...((..(((..(((((-.....(((((((((((.((....)).)))))))))))...))))..)..)))......))....)))) ( -27.80, z-score = -2.31, R) >dp4.chr4_group3 9730799 103 + 11692001 AGUAGGGACAAGAAAGGACCACAGCUGCAGCUGACAAAACUGAAAUUAAAGAAGGAGCUAAAGGGGU----UUAAGGAAAUAUUAUAGUAUAAAGAAAAAGAAGGAU .((.((..(......)..)).((((....))))))...((((.(((........(((((.....)))----))........))).)))).................. ( -14.29, z-score = -1.36, R) >consensus ________________GGCAGAGACUGCAGCUGACAAA_CUCAAAUUGAAGGGGGCACAAAAGUUUUCCCUUCAGUGCAUUUGGAUUGGCUAAAUAAACGGAGCUGC ..........................((((((............(((((((((((.((....)).)))))))))))..((((((.....))))))......)))))) (-16.70 = -18.70 + 2.00)
| Location | 1,262,236 – 1,262,326 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 77.91 |
| Shannon entropy | 0.38377 |
| G+C content | 0.45061 |
| Mean single sequence MFE | -19.93 |
| Consensus MFE | -13.32 |
| Energy contribution | -13.88 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.676487 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
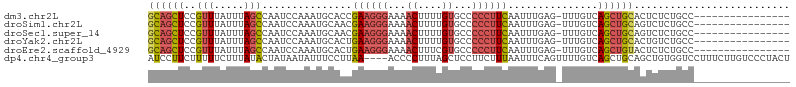
>dm3.chr2L 1262236 90 - 23011544 GCAGCUCCGUUUAUUUAGCCAAUCCAAAUGCACCGAAGGGAAAACUUUUGUGCCCCCUUCAAUUUGAG-UUUGUCAGCUGCACUCUCUGCC---------------- ((((((..(((.....)))(((.((((((.....((((((...((....))...)))))).))))).)-.)))..))))))..........---------------- ( -22.30, z-score = -1.77, R) >droSim1.chr2L 1234252 90 - 22036055 GCAGCUCCGUUUAUUUAGCCAAUCCAAAUGCAACGAAGGGAAAACUUUUGUGCCCCCUUCAAUUUGAG-UUUGUCAGCUGCAGUCUCUGCC---------------- ((((((..(((.....))).....(((((.(((.((((((...((....))...))))))...))).)-))))..))))))..........---------------- ( -23.20, z-score = -1.78, R) >droSec1.super_14 1212827 90 - 2068291 GCAGCUCCGUUUAUUUAGCCAAUCCAAAUGCAACGAAGGGAAAACUUUUGUGCCCCCUUCAAUUUGAG-UUUGUCAGCUGCAGUCUCUGCC---------------- ((((((..(((.....))).....(((((.(((.((((((...((....))...))))))...))).)-))))..))))))..........---------------- ( -23.20, z-score = -1.78, R) >droYak2.chr2L 1237346 90 - 22324452 GCAGCUCCGUUUAUUUAGCCAAUCCAAAUGCACUGAAGGGAAAACUUUUGUGCCCCCUUCAAUUUGAG-UUUGUCAGCUGCACUGUCUGCC---------------- ((((((..(((.....))).....(((((.((.(((((((...((....))...)))))))...)).)-))))..))))))..........---------------- ( -23.70, z-score = -1.77, R) >droEre2.scaffold_4929 1302851 90 - 26641161 GCAGCUCCGUUUAUUUAGCCAAUCCAAAUGCACUGAAGGGAAAACUUUCGUGCCCCCUUCAAUUUGAG-UUUGUCAGCUGUACUCUCUGCC---------------- ((((((..(((.....))).....(((((.((.(((((((...((....))...)))))))...)).)-))))..))))))..........---------------- ( -20.70, z-score = -1.51, R) >dp4.chr4_group3 9730799 103 - 11692001 AUCCUUCUUUUUCUUUAUACUAUAAUAUUUCCUUAA----ACCCCUUUAGCUCCUUCUUUAAUUUCAGUUUUGUCAGCUGCAGCUGUGGUCCUUUCUUGUCCCUACU ....................................----..................................((((....)))).((..(......)..)).... ( -6.50, z-score = 0.22, R) >consensus GCAGCUCCGUUUAUUUAGCCAAUCCAAAUGCACCGAAGGGAAAACUUUUGUGCCCCCUUCAAUUUGAG_UUUGUCAGCUGCACUCUCUGCC________________ ((((((..(((.....)))...............((((((...((....))...))))))...............)))))).......................... (-13.32 = -13.88 + 0.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:08:06 2011