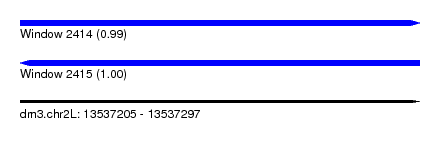
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 13,537,205 – 13,537,297 |
| Length | 92 |
| Max. P | 0.999632 |

| Location | 13,537,205 – 13,537,297 |
|---|---|
| Length | 92 |
| Sequences | 7 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 66.27 |
| Shannon entropy | 0.62949 |
| G+C content | 0.44517 |
| Mean single sequence MFE | -32.91 |
| Consensus MFE | -12.12 |
| Energy contribution | -13.19 |
| Covariance contribution | 1.07 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.58 |
| SVM RNA-class probability | 0.992998 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
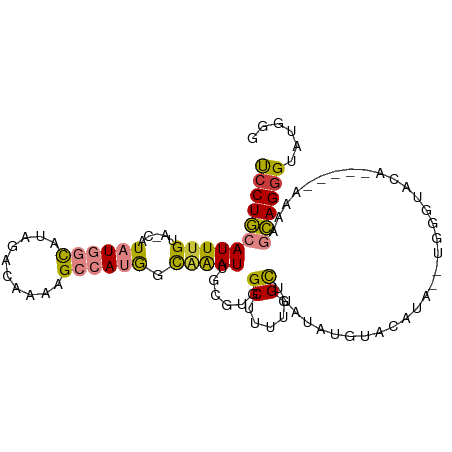
>dm3.chr2L 13537205 92 + 23011544 UCCUGCAUUUGUA-CAUAUGGCAUAGACAAAAGCCAUGGCAAAUUGCGUGGCUUUUGGCUUAUAUGUACAUA--UGGUUA--------UGCGCAGGGUAUGGG ((((((.......-(((((((((((..(((((((((((........))))))))))).....))))).))))--))....--------...))))))...... ( -35.24, z-score = -3.30, R) >droPer1.super_5 3218907 91 + 6813705 CCCUAAAUUUGUAUCAUAUCUUACC-ACCCUGGCCAUAGUUGAUGUCAUAGCGAA-GGUAUCCUUUUCCGUGAAUGAGGCAA-----AAG--UAGGGGAG--- (((((..(((((.((((......((-.....))............(((..(.(((-((...))))).)..))))))).))))-----)..--)))))...--- ( -22.70, z-score = -0.87, R) >dp4.chr4_group1 3269721 95 - 5278887 CCCUAAAUUUGUAUCAUAUCUUACC-ACCCCGGCCAUAGUGGAUGUCAUAGCGAA-GGUAUCCUUUUCCGUGCAUGAGGCAA-----GAGAGUAGGGGGAGG- (((((..(((((.((........((-((..........))))(((.((..(.(((-((...))))).)..))))))).))))-----)....))))).....- ( -22.40, z-score = 0.94, R) >droEre2.scaffold_4929 14726811 86 - 26641161 UCCUGCAUUUGCA-CAUAUGGCAUAGACAAAAGCCAUGGCAAAUUGCGUGGCUUUUGGCUUAUA----CAUA--UGGGGA----------UGCAGGGUAUGGG ((((((((((...-((((((....((.(((((((((((........))))))))))).))....----))))--)).)))----------)))))))...... ( -37.70, z-score = -5.02, R) >droYak2.chr2L 9961154 102 + 22324452 UCCUGCAUUUGUA-CAUAUGGCAUAGACAAAAGCCAUGGCAGAUUGCGUCGCUUUUGGCUUAUAUGUAUAUACAUAUGUACAUAUGGGAAUGCAGAGUAUGGG ..(((((((((((-((((((..(((.(((.((((((.(((.(((...))))))..))))))...))).))).))))))))))......)))))))........ ( -33.50, z-score = -2.52, R) >droSec1.super_16 1677124 100 + 1878335 UCCUGCAUUUGUA-CAUAUGGCAUAGACAAAAGCCAGGCCAAAUUGCGUGGCUUUUGGCUUAUAUGUACAUA--UGGGUACAUGGGUAUAAGCAGGGUAUGGG ((((((..(((((-(.((((.......(((((((((.((......)).)))))))))((((((((....)))--))))).)))).))))))))))))...... ( -39.40, z-score = -3.89, R) >droSim1.chr2L 13310224 100 + 22036055 UCCUGCAUUUGUA-CAUAUGGCAUAGACAAAAGCCAGGCCAAAUUGCGUGGCUUUUGGCUUAUAUGUACAUA--UGGGUACAUGGGUAUAAGCAGGGUAUGGG ((((((..(((((-(.((((.......(((((((((.((......)).)))))))))((((((((....)))--))))).)))).))))))))))))...... ( -39.40, z-score = -3.89, R) >consensus UCCUGCAUUUGUA_CAUAUGGCAUAGACAAAAGCCAUGGCAAAUUGCGUGGCUUUUGGCUUAUAUGUACAUA__UGGGUACA_____AAAAGCAGGGUAUGGG ((((((...(((........)))....(((((((.(((........))).)))))))..................................))))))...... (-12.12 = -13.19 + 1.07)

| Location | 13,537,205 – 13,537,297 |
|---|---|
| Length | 92 |
| Sequences | 7 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 66.27 |
| Shannon entropy | 0.62949 |
| G+C content | 0.44517 |
| Mean single sequence MFE | -29.46 |
| Consensus MFE | -13.34 |
| Energy contribution | -12.24 |
| Covariance contribution | -1.10 |
| Combinations/Pair | 1.50 |
| Mean z-score | -3.22 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.11 |
| SVM RNA-class probability | 0.999632 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
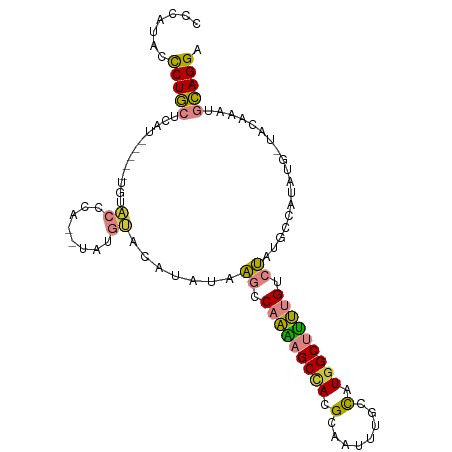
>dm3.chr2L 13537205 92 - 23011544 CCCAUACCCUGCGCA--------UAACCA--UAUGUACAUAUAAGCCAAAAGCCACGCAAUUUGCCAUGGCUUUUGUCUAUGCCAUAUG-UACAAAUGCAGGA .......((((((..--------....).--..(((((((((.((.(((((((((.((.....))..))))))))).)).....)))))-))))...))))). ( -29.30, z-score = -3.74, R) >droPer1.super_5 3218907 91 - 6813705 ---CUCCCCUA--CUU-----UUGCCUCAUUCACGGAAAAGGAUACC-UUCGCUAUGACAUCAACUAUGGCCAGGGU-GGUAAGAUAUGAUACAAAUUUAGGG ---...(((((--..(-----(((..((((((.(......)..((((-((.((((((........)))))).)))))-)....)).))))..))))..))))) ( -24.40, z-score = -1.69, R) >dp4.chr4_group1 3269721 95 + 5278887 -CCUCCCCCUACUCUC-----UUGCCUCAUGCACGGAAAAGGAUACC-UUCGCUAUGACAUCCACUAUGGCCGGGGU-GGUAAGAUAUGAUACAAAUUUAGGG -.....(((((...((-----(((((.......(......)...(((-(..((((((........))))))..))))-)))))))...(((....)))))))) ( -21.70, z-score = 0.57, R) >droEre2.scaffold_4929 14726811 86 + 26641161 CCCAUACCCUGCA----------UCCCCA--UAUG----UAUAAGCCAAAAGCCACGCAAUUUGCCAUGGCUUUUGUCUAUGCCAUAUG-UGCAAAUGCAGGA .......((((((----------(..(((--((((----((((.(.(((((((((.((.....))..))))))))).))))).))))))-.)...))))))). ( -32.00, z-score = -4.91, R) >droYak2.chr2L 9961154 102 - 22324452 CCCAUACUCUGCAUUCCCAUAUGUACAUAUGUAUAUACAUAUAAGCCAAAAGCGACGCAAUCUGCCAUGGCUUUUGUCUAUGCCAUAUG-UACAAAUGCAGGA .......((((((((......((((((((((((((.(((...((((((...(((........)))..)))))).))).)))).))))))-)))))))))))). ( -34.20, z-score = -4.71, R) >droSec1.super_16 1677124 100 - 1878335 CCCAUACCCUGCUUAUACCCAUGUACCCA--UAUGUACAUAUAAGCCAAAAGCCACGCAAUUUGGCCUGGCUUUUGUCUAUGCCAUAUG-UACAAAUGCAGGA .......(((((........(((....))--).(((((((((.((.(((((((((.((......)).))))))))).)).....)))))-))))...))))). ( -32.30, z-score = -4.04, R) >droSim1.chr2L 13310224 100 - 22036055 CCCAUACCCUGCUUAUACCCAUGUACCCA--UAUGUACAUAUAAGCCAAAAGCCACGCAAUUUGGCCUGGCUUUUGUCUAUGCCAUAUG-UACAAAUGCAGGA .......(((((........(((....))--).(((((((((.((.(((((((((.((......)).))))))))).)).....)))))-))))...))))). ( -32.30, z-score = -4.04, R) >consensus CCCAUACCCUGCUCAU_____UGUACCCA__UAUGUACAUAUAAGCCAAAAGCCACGCAAUUUGCCAUGGCUUUUGUCUAUGCCAUAUG_UACAAAUGCAGGA .......(((((...............(......)........((.(((((((((.(........).))))))))).))..................))))). (-13.34 = -12.24 + -1.10)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:38:13 2011