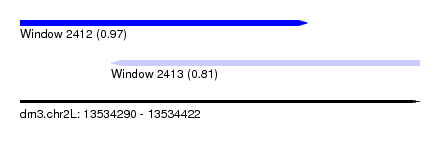
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 13,534,290 – 13,534,422 |
| Length | 132 |
| Max. P | 0.970907 |

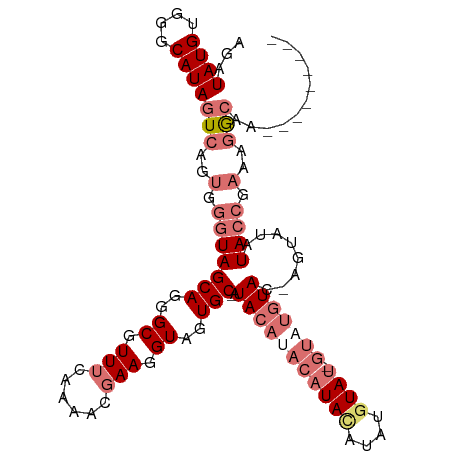
| Location | 13,534,290 – 13,534,385 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 74.79 |
| Shannon entropy | 0.40619 |
| G+C content | 0.42638 |
| Mean single sequence MFE | -27.88 |
| Consensus MFE | -10.98 |
| Energy contribution | -10.73 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.29 |
| Mean z-score | -3.20 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.84 |
| SVM RNA-class probability | 0.970907 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 13534290 95 + 23011544 ACUCAGCAGAGGGUACCCGCCGUGGCUCCUUUG--CCUUUCGGUAUAUAUUGUACAUAC--------AUAUGUAUGUAUGAAUGUAUUGCACUACCUUCGUUUUG ........(((((((...((((.(((......)--))...))))(((((((((((((((--------....)))))))).))))))).....)))))))...... ( -31.60, z-score = -3.70, R) >droSim1.chr2L 13307494 94 + 22036055 ACUCAGCAGAGGGGACCCACCGUGGCUCCUUUG--CCUUUCGGUAUAU---ACAUACAUAU-----UGUACAUAUGUAUGUUUGUAU-GCACUACCUUCGUUUUG .....(((((((((.((......))))))))))--)......((((((---((((((((((-----.....))))))))))..))))-))............... ( -29.90, z-score = -3.30, R) >droYak2.chr2L 9958221 79 + 22324452 ACUCAGCAGAGGG------------CUCCUUU---CCUUUCGCUAUAUUGUAUGUAC----------AUACAUAUGUAUGUGUGUAU-GCACUACCUUCGUUUUG ....(((((((((------------......)---))))).))).....((..((((----------((((((....))))))))))-..))............. ( -19.80, z-score = -2.17, R) >droEre2.scaffold_4929 14720650 104 - 26641161 ACUCAGCAGAGGGCACCCACCGUGGCUCCUUUGCUCCUUUCGUUAUAUUGUACAUACUCGUACAUACAUACAUACAUAUGUAUGUAU-GCAGUACCUUCGUUUUG ....((((((((((((.....)).)).))))))))...................((((((((((((((((......)))))))))))-).))))........... ( -30.20, z-score = -3.62, R) >consensus ACUCAGCAGAGGG_ACCCACCGUGGCUCCUUUG__CCUUUCGGUAUAUUGUACAUACAC________AUACAUAUGUAUGUAUGUAU_GCACUACCUUCGUUUUG .....((.(((((..((......))..)))))...................................((((((((....)))))))).))............... (-10.98 = -10.73 + -0.25)

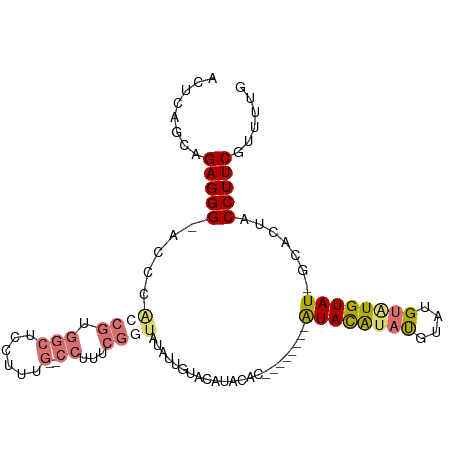
| Location | 13,534,320 – 13,534,422 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 81.15 |
| Shannon entropy | 0.29219 |
| G+C content | 0.39819 |
| Mean single sequence MFE | -27.98 |
| Consensus MFE | -12.85 |
| Energy contribution | -13.35 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.85 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.814732 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 13534320 102 - 23011544 AGAUAUGUGGGCAUAGUCAGUGGGUAGCAGGGCGUUUCAAAACGAAGGUAGUGCAAUACAUUCAUACAUACAUAU-GUAUGUAC-AAUAUAUACCGAAAGGCAA--------- ...((((....))))(((..(.(((((((..((.((((.....))))))..)))..........(((((((....-))))))).-......)))).)..)))..--------- ( -24.30, z-score = -1.78, R) >droSim1.chr2L 13307524 101 - 22036055 AGAUAUGUGGGCAUAGUCAGUGGGUAGCAGGGCGUUUCAAAACGAAGGUAGUGC-AUACAAACAUACAUAUGUACAAUAUGUAU--GUAUAUACCGAAAGGCAA--------- ...((((....))))(((..(.(((((((..((.((((.....))))))..)))-......((((((((((.....))))))))--))...)))).)..)))..--------- ( -28.40, z-score = -3.11, R) >droYak2.chr2L 9958239 98 - 22324452 AGAUAUGUGGGCAUAGUCAGUGGGUAGCAGGGCGUUUCAAAACGAAGGUAGUGC-AUACACACAUACAUAUGUAU-GUACAUAC-A--AUAUAGCGAAAGGAA---------- ...((((((.(((((((..(((.((((((..((.((((.....))))))..)))-.))).)))..)).)))))..-.)))))).-.--......(....)...---------- ( -22.10, z-score = -1.65, R) >droEre2.scaffold_4929 14720680 111 + 26641161 AGAUAUGUGGGCAUAGUCAGUGGGUAGCAGGGCGUUACAAAACGAAGGUACUGC-AUACAUACAUAUGUAUGUAU-GUAUGUACGAGUAUGUACAAUAUAACGAAAGGAGCAA ...(((((..(((((.((.((..(((((.....)))))...))....(((((((-((((((((....))))))))-))).)))))).)))))...))))).(....)...... ( -37.10, z-score = -4.85, R) >consensus AGAUAUGUGGGCAUAGUCAGUGGGUAGCAGGGCGUUUCAAAACGAAGGUAGUGC_AUACAUACAUACAUAUGUAU_GUAUGUAC_AGUAUAUACCGAAAGGCAA_________ .((((((....))).))).(((.((((((..((.(((......))).))..)))..))).))).(((((((.....))))))).............................. (-12.85 = -13.35 + 0.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:38:12 2011