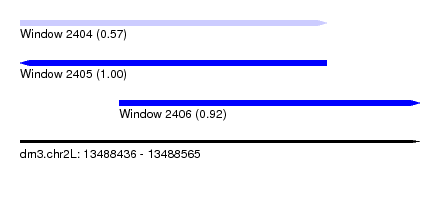
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 13,488,436 – 13,488,565 |
| Length | 129 |
| Max. P | 0.999820 |

| Location | 13,488,436 – 13,488,535 |
|---|---|
| Length | 99 |
| Sequences | 10 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 75.37 |
| Shannon entropy | 0.50118 |
| G+C content | 0.39819 |
| Mean single sequence MFE | -23.13 |
| Consensus MFE | -5.09 |
| Energy contribution | -6.09 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.84 |
| Structure conservation index | 0.22 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.565440 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 13488436 99 + 23011544 -----CAAAAACGGCAAAAUGCAAAUAAAAAAA--AUUUUCCCUUUGACAAUGCCGACCGAUU------CGCGUCGAGUCGACAGCCAAAGUGAAACUUUUGCCUUAAAUUG -----.......((((((((....)).......--..((((.(((((....((.((((((((.------...)))).)))).))..))))).))))..))))))........ ( -21.60, z-score = -2.31, R) >droSim1.chr2L 13270317 99 + 22036055 -----CAAAAACGGCAAAAUGCAAAUAAAAAAA--AUUUUCCCUUUGACAAUGCCGACCGAUU------CGCGUCGAGUCGACAGCCAAAGUGAAACUUUUGCCUUAAAUUG -----.......((((((((....)).......--..((((.(((((....((.((((((((.------...)))).)))).))..))))).))))..))))))........ ( -21.60, z-score = -2.31, R) >droSec1.super_16 1633692 99 + 1878335 -----CAAAAACGGCAAAAUGCAAAUAAAAAAA--AUUUUCCCUUUGACAAUGCCGACCGAUU------CGCGUCGAGUCGACAGCCAAAGUGAAACUUUUGCCUUAAAUUG -----.......((((((((....)).......--..((((.(((((....((.((((((((.------...)))).)))).))..))))).))))..))))))........ ( -21.60, z-score = -2.31, R) >droYak2.chr2L 9916630 101 + 22324452 -----CAAAAACGGCAAAAUGCAAAUAAAAAAAAUUGUUUCCCUUUGACAAUGCCGACCGAUU------CGCGUCGAGUCGACAGCCAAAGUGAAACUUUUGCCUUAAAUUG -----.......(((((((.................(((((.(((((....((.((((((((.------...)))).)))).))..))))).))))))))))))........ ( -25.22, z-score = -3.18, R) >droEre2.scaffold_4929 14680274 99 - 26641161 -----CAAAAACGGCAAAAUGCAAAUAAAAAAA--UGUUUCCCUUUGACAAUGCCGACCGAUU------CGCGUCGAGUCGACAGCCAAAGUGAAACUUUUGCCUUAAAUUG -----.......(((((((..............--.(((((.(((((....((.((((((((.------...)))).)))).))..))))).))))))))))))........ ( -25.36, z-score = -3.33, R) >droAna3.scaffold_12943 2192851 99 + 5039921 -----CAAAAACGGCAAAAUGAAAGUGAAACAA--AUUUUCCCUUUGACAAUGCCGACCGUUU------CUAGUCGUGUCGGCUGCCAAAGUGAAAACUUUGCCUCAAAUUG -----.......((((((((....)).......--.(((((.(((((.((..((((((((...------.....)).)))))))).))))).))))).))))))........ ( -27.80, z-score = -3.68, R) >dp4.chr4_group1 3214169 105 - 5278887 -----CAAAAACGGCAAACUGAAAACGAAACAA--AUUUUCACUUUGACAAUACCAACCCGAUUCGAGUCGAGUCGCGUCGGCUGCCAAAGCGAAAACUUUGCCUCAAAUUU -----.......((((((.(....)........--.(((((.(((((.((...((.((.(((((((...))))))).)).)).)).))))).))))).))))))........ ( -24.90, z-score = -2.28, R) >droPer1.super_5 3162643 105 + 6813705 -----CAAAAACGGCAAACUGAAAACGAAACAA--AUUUUCACUUUGACAAUACCAACCCGAUUCCAGUCGAGUCGCGUCGGCUGCCAAAGCGAAAACUUUGCCUCAAAUUU -----.......((((((.(....)........--.(((((.(((((.((...((.((.((((((.....)))))).)).)).)).))))).))))).))))))........ ( -23.50, z-score = -2.25, R) >droWil1.scaffold_180708 7322031 91 - 12563649 CAAAAGAGAAAUGCGAAAAUGCAACUAACGAGA--AUGUUCACUUUGACAAUGCCC-CCCGAC------------------AAUGCCAAAGUGAAAACUUUGCCUCGAAUUG .....(((...(((......))).....((((.--...(((((((((.((.((...-.....)------------------).)).)))))))))...)))).)))...... ( -17.40, z-score = -3.05, R) >droMoj3.scaffold_6500 3335160 90 - 32352404 -----CAAAAA--GCAUAAGAAAUACAAAAUA---CUUUUCGCUUUGACA-UUUCGAUUGGGG------AAUGUCAAAGAGA-----AAAUGGAAUAUUUUGACCCAAAUUG -----......--............(((((((---((((((.((((((((-((((......))------)))))))))).))-----))).)...))))))).......... ( -22.30, z-score = -3.75, R) >consensus _____CAAAAACGGCAAAAUGCAAAUAAAAAAA__AUUUUCCCUUUGACAAUGCCGACCGAUU______CGCGUCGAGUCGACAGCCAAAGUGAAAAUUUUGCCUCAAAUUG ............((((((..................(((((.(((((.......................................))))).))))).))))))........ ( -5.09 = -6.09 + 1.00)

| Location | 13,488,436 – 13,488,535 |
|---|---|
| Length | 99 |
| Sequences | 10 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 75.37 |
| Shannon entropy | 0.50118 |
| G+C content | 0.39819 |
| Mean single sequence MFE | -31.95 |
| Consensus MFE | -17.64 |
| Energy contribution | -18.94 |
| Covariance contribution | 1.30 |
| Combinations/Pair | 1.28 |
| Mean z-score | -3.95 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.48 |
| SVM RNA-class probability | 0.999820 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 13488436 99 - 23011544 CAAUUUAAGGCAAAAGUUUCACUUUGGCUGUCGACUCGACGCG------AAUCGGUCGGCAUUGUCAAAGGGAAAAU--UUUUUUUAUUUGCAUUUUGCCGUUUUUG----- ........(((((((.((((.((((((((((((((.(((....------..))))))))))..))))))).))))..--..............))))))).......----- ( -30.73, z-score = -3.46, R) >droSim1.chr2L 13270317 99 - 22036055 CAAUUUAAGGCAAAAGUUUCACUUUGGCUGUCGACUCGACGCG------AAUCGGUCGGCAUUGUCAAAGGGAAAAU--UUUUUUUAUUUGCAUUUUGCCGUUUUUG----- ........(((((((.((((.((((((((((((((.(((....------..))))))))))..))))))).))))..--..............))))))).......----- ( -30.73, z-score = -3.46, R) >droSec1.super_16 1633692 99 - 1878335 CAAUUUAAGGCAAAAGUUUCACUUUGGCUGUCGACUCGACGCG------AAUCGGUCGGCAUUGUCAAAGGGAAAAU--UUUUUUUAUUUGCAUUUUGCCGUUUUUG----- ........(((((((.((((.((((((((((((((.(((....------..))))))))))..))))))).))))..--..............))))))).......----- ( -30.73, z-score = -3.46, R) >droYak2.chr2L 9916630 101 - 22324452 CAAUUUAAGGCAAAAGUUUCACUUUGGCUGUCGACUCGACGCG------AAUCGGUCGGCAUUGUCAAAGGGAAACAAUUUUUUUUAUUUGCAUUUUGCCGUUUUUG----- ........((((((((((((.((((((((((((((.(((....------..))))))))))..))))))).)))))((......)).......))))))).......----- ( -33.20, z-score = -4.21, R) >droEre2.scaffold_4929 14680274 99 + 26641161 CAAUUUAAGGCAAAAGUUUCACUUUGGCUGUCGACUCGACGCG------AAUCGGUCGGCAUUGUCAAAGGGAAACA--UUUUUUUAUUUGCAUUUUGCCGUUUUUG----- ........((((((((((((.((((((((((((((.(((....------..))))))))))..))))))).)))))(--(......)).....))))))).......----- ( -32.50, z-score = -4.08, R) >droAna3.scaffold_12943 2192851 99 - 5039921 CAAUUUGAGGCAAAGUUUUCACUUUGGCAGCCGACACGACUAG------AAACGGUCGGCAUUGUCAAAGGGAAAAU--UUGUUUCACUUUCAUUUUGCCGUUUUUG----- .....(((((((((.(((((.((((((((((((((.((.....------...))))))))..))))))))))))).)--))))))))....................----- ( -34.50, z-score = -4.51, R) >dp4.chr4_group1 3214169 105 + 5278887 AAAUUUGAGGCAAAGUUUUCGCUUUGGCAGCCGACGCGACUCGACUCGAAUCGGGUUGGUAUUGUCAAAGUGAAAAU--UUGUUUCGUUUUCAGUUUGCCGUUUUUG----- ......((((((((.((((((((((((((((((((.(((.(((...))).))).))))))..)))))))))))))))--))))))).....................----- ( -40.80, z-score = -4.67, R) >droPer1.super_5 3162643 105 - 6813705 AAAUUUGAGGCAAAGUUUUCGCUUUGGCAGCCGACGCGACUCGACUGGAAUCGGGUUGGUAUUGUCAAAGUGAAAAU--UUGUUUCGUUUUCAGUUUGCCGUUUUUG----- ......((((((((.((((((((((((((((((((.(((.((.....)).))).))))))..)))))))))))))))--))))))).....................----- ( -40.10, z-score = -4.45, R) >droWil1.scaffold_180708 7322031 91 + 12563649 CAAUUCGAGGCAAAGUUUUCACUUUGGCAUU------------------GUCGGG-GGGCAUUGUCAAAGUGAACAU--UCUCGUUAGUUGCAUUUUCGCAUUUCUCUUUUG .....(((((....((.((((((((((((.(------------------(((...-.)))).)))))))))))).))--))))).....(((......)))........... ( -26.70, z-score = -3.85, R) >droMoj3.scaffold_6500 3335160 90 + 32352404 CAAUUUGGGUCAAAAUAUUCCAUUU-----UCUCUUUGACAUU------CCCCAAUCGAAA-UGUCAAAGCGAAAAG---UAUUUUGUAUUUCUUAUGC--UUUUUG----- .....((((.((((((((....(((-----((.((((((((((------.(......).))-)))))))).))))))---))))))).....))))...--......----- ( -19.50, z-score = -3.38, R) >consensus CAAUUUAAGGCAAAAGUUUCACUUUGGCUGUCGACUCGACGCG______AAUCGGUCGGCAUUGUCAAAGGGAAAAU__UUUUUUUAUUUGCAUUUUGCCGUUUUUG_____ ........((((((((((((.(((((((.((((((...................))))))...))))))).))))).................)))))))............ (-17.64 = -18.94 + 1.30)
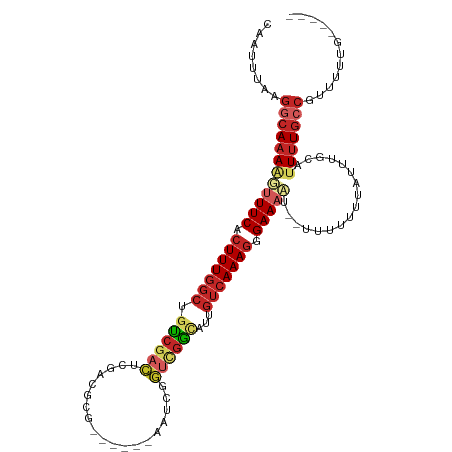
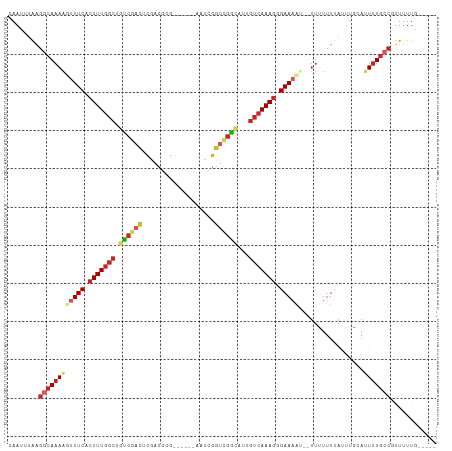
| Location | 13,488,468 – 13,488,565 |
|---|---|
| Length | 97 |
| Sequences | 11 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 76.32 |
| Shannon entropy | 0.47121 |
| G+C content | 0.43175 |
| Mean single sequence MFE | -23.46 |
| Consensus MFE | -8.21 |
| Energy contribution | -8.02 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.80 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.924164 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 13488468 97 + 23011544 UCCCUUUGACAAUGCCGACC-GAUUCGCGUCGAGUCG-----ACAGCCAAAGUGAAACUUUUGCCUUAAAUUGAAAUGCAC-AUUUUAAUUUCGGCCCGAAACC--------- ((.(((((....((.(((((-(((....)))).))))-----.))..))))).)).......(((..((((((((((....-)))))))))).)))........--------- ( -23.60, z-score = -2.60, R) >droSim1.chr2L 13270349 97 + 22036055 UCCCUUUGACAAUGCCGACC-GAUUCGCGUCGAGUCG-----ACAGCCAAAGUGAAACUUUUGCCUUAAAUUGAAAUGCAC-AUUUUAAUUUCGGCCCGAAACC--------- ((.(((((....((.(((((-(((....)))).))))-----.))..))))).)).......(((..((((((((((....-)))))))))).)))........--------- ( -23.60, z-score = -2.60, R) >droSec1.super_16 1633724 97 + 1878335 UCCCUUUGACAAUGCCGACC-GAUUCGCGUCGAGUCG-----ACAGCCAAAGUGAAACUUUUGCCUUAAAUUGAAAUGCAC-AUUUUAAUUUCGGCCCAAAACC--------- ((.(((((....((.(((((-(((....)))).))))-----.))..))))).)).......(((..((((((((((....-)))))))))).)))........--------- ( -23.60, z-score = -2.88, R) >droYak2.chr2L 9916664 97 + 22324452 UCCCUUUGACAAUGCCGACC-GAUUCGCGUCGAGUCG-----ACAGCCAAAGUGAAACUUUUGCCUUAAAUUGAAAUGCAC-AUUUUAAUUUCGGCCCGAAACC--------- ((.(((((....((.(((((-(((....)))).))))-----.))..))))).)).......(((..((((((((((....-)))))))))).)))........--------- ( -23.60, z-score = -2.60, R) >droEre2.scaffold_4929 14680306 97 - 26641161 UCCCUUUGACAAUGCCGACC-GAUUCGCGUCGAGUCG-----ACAGCCAAAGUGAAACUUUUGCCUUAAAUUGAAAUGCAC-AUUUUAAUUUCGGCCCGAAACC--------- ((.(((((....((.(((((-(((....)))).))))-----.))..))))).)).......(((..((((((((((....-)))))))))).)))........--------- ( -23.60, z-score = -2.60, R) >droAna3.scaffold_12943 2192883 97 + 5039921 UCCCUUUGACAAUGCCGACC-GUUUCUAGUCGUGUCG-----GCUGCCAAAGUGAAAACUUUGCCUCAAAUUGAAAUGCAC-AUUUUAAUUUCGGCCCGAAACC--------- ((.(((((.((..(((((((-(........)).))))-----)))).))))).)).......(((..((((((((((....-)))))))))).)))........--------- ( -27.50, z-score = -3.62, R) >dp4.chr4_group1 3214201 103 - 5278887 UCACUUUGACAAUACCAACCCGAUUCGAGUCGAGUCGCGUCGGCUGCCAAAGCGAAAACUUUGCCUCAAAUUUAAAUGCAC-AUUUUAAUUUCGGCCCGAAACC--------- ((.(((((.((...((.((.(((((((...))))))).)).)).)).))))).)).......(((..(((((.((((....-)))).))))).)))........--------- ( -22.50, z-score = -1.42, R) >droPer1.super_5 3162675 103 + 6813705 UCACUUUGACAAUACCAACCCGAUUCCAGUCGAGUCGCGUCGGCUGCCAAAGCGAAAACUUUGCCUCAAAUUUAAAUGCAC-AUUUUAAUUUCGGCCCGAAACC--------- ((.(((((.((...((.((.((((((.....)))))).)).)).)).))))).)).......(((..(((((.((((....-)))).))))).)))........--------- ( -21.10, z-score = -1.40, R) >droWil1.scaffold_180708 7322068 93 - 12563649 UCACUUUGACAAUGC-----CCCCC---GACAAUGCC----------AAAGUGAAA-ACUUUGCCUCGAAUUGAAAUGAAA-AUUUUAAUUUCGGCUCAAAACCGUUUUAUAU ((((((((.((.((.-----.....---..)).)).)----------)))))))..-.....(((..((((((((((....-)))))))))).)))................. ( -22.50, z-score = -4.67, R) >droVir3.scaffold_12723 5318683 90 - 5802038 UCGCUUUGACAAUCCAAAUG-GGCC---GCCAAAGCG----------AAAACAAAAUACUUUGCCACAAAUUGAAAUGCAGCAUUUUAAUUUCGGUUCGAAAAG--------- ((((((((.(..(((....)-))..---).)))))))----------)..............(((..(((((((((((...))))))))))).)))........--------- ( -21.00, z-score = -1.89, R) >droGri2.scaffold_15252 13118902 89 - 17193109 UCGCUUUGACAACUCGAAAACGGUU---GCCAAAGCG----------GAAU--AAAUACUUUGCCCAAAAUUGAAAUGCACCAUUUUAAUUUCGGAUCAAAAAG--------- ((((((((.((((.((....)))))---).)))))))----------)...--......((((.((.(((((((((((...))))))))))).))..))))...--------- ( -25.50, z-score = -4.50, R) >consensus UCCCUUUGACAAUGCCGACC_GAUUCGCGUCGAGUCG_____ACAGCCAAAGUGAAAACUUUGCCUCAAAUUGAAAUGCAC_AUUUUAAUUUCGGCCCGAAACC_________ ....((((.............(((....)))...............................(((..((((((((((.....)))))))))).))).))))............ ( -8.21 = -8.02 + -0.19)

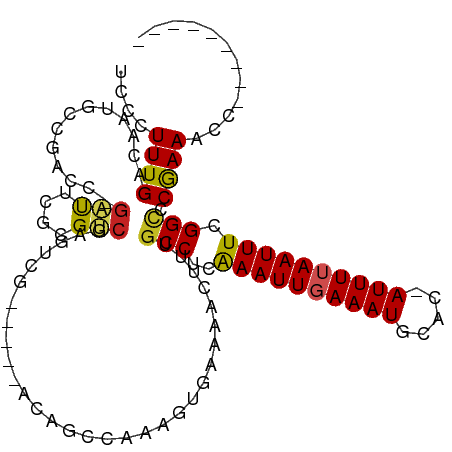
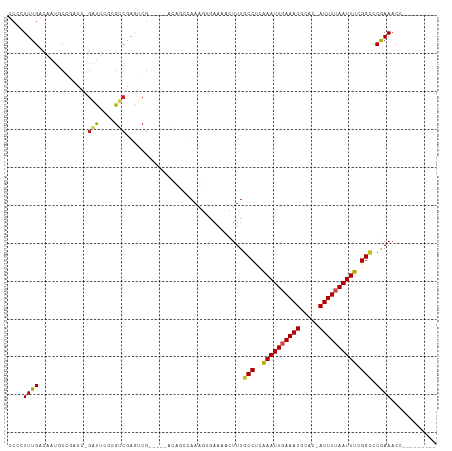
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:38:06 2011