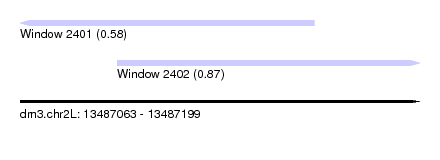
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 13,487,063 – 13,487,199 |
| Length | 136 |
| Max. P | 0.867931 |

| Location | 13,487,063 – 13,487,163 |
|---|---|
| Length | 100 |
| Sequences | 11 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 70.99 |
| Shannon entropy | 0.60463 |
| G+C content | 0.40018 |
| Mean single sequence MFE | -21.64 |
| Consensus MFE | -7.76 |
| Energy contribution | -8.30 |
| Covariance contribution | 0.54 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.577509 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
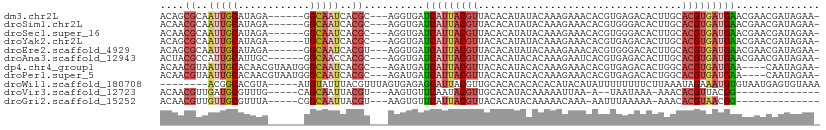
>dm3.chr2L 13487063 100 - 23011544 ACAGCGCAAUUGCAUAGA------GGCAAUCACGC---AGGUGAUCAUUACGUUACACAUAUACAAAGAAACACGUGAGACACUUGCACGUGAUGAACGAACGAUAGAA- .((.(((.(((((.....------.)))))...((---(((((.((.((((((...................)))))))))))))))..))).))..............- ( -22.91, z-score = -1.91, R) >droSim1.chr2L 13268946 100 - 22036055 ACAACGCAAUUGCAUAGA------GGCAAUCACGC---AGGUGAUCAUUACGUUACACAUAUACAAAGAAACACGUGGGACACUUGCACGUGAUGAACGAACGAUAGAA- .((.(((.(((((.....------.)))))...((---(((((.((.((((((...................)))))))))))))))..))).))..............- ( -22.11, z-score = -1.86, R) >droSec1.super_16 1632321 100 - 1878335 ACAACGCAAUUGCAUAGA------GGCAAUCACGC---AGGUGAUCAUUACGUUACACAUAUACAAAGAAACACGUGGGACACUUGCACGUGAUGAACGAACGAUAGAA- .((.(((.(((((.....------.)))))...((---(((((.((.((((((...................)))))))))))))))..))).))..............- ( -22.11, z-score = -1.86, R) >droYak2.chr2L 9915237 100 - 22324452 ACAGCGCAAUUGCAUAGA------UGCAAUCACGC---AGGUGAUCAUUACGUUACACAUAUACAAAGAAACACGUGAGACACUUGCACGUGAUGAACGAACGAUAGAA- .((.(((.((((((....------))))))...((---(((((.((.((((((...................)))))))))))))))..))).))..............- ( -23.71, z-score = -1.96, R) >droEre2.scaffold_4929 14678879 100 + 26641161 ACAGCGCAAUUGCAUAGA------GGCAAUCACGU---AGGUGAUCAUUACGUUACACAUAUACAAAGAAACACGUGGGACACUUGCACGUGAUGAACGAACGAUAGAA- .((.(((.(((((.....------.)))))...((---(((((.((.((((((...................)))))))))))))))..))).))..............- ( -20.11, z-score = -0.62, R) >droAna3.scaffold_12943 2191329 100 - 5039921 ACUACGCCAUUGCAUUGC------GGCAACCACGC---AGGUGAUCAUUACGUUACACAUACACAAAGAAUCACGUGAGACACUUGCACGUGAUGAACGAACGAUAGAA- .(((((......(((..(------(....)...((---(((((.((.((((((...................)))))))))))))))..)..)))......)).)))..- ( -24.71, z-score = -1.88, R) >dp4.chr4_group1 3212715 102 + 5278887 ACAACGUAAUUGCACAACGUAAUGGGCAAUCACGC---AGAUGAUCAUUACGUUACACAUACACAAAGAAACACGUGAGACACUGGCACGUGAUGAA----CAAUAGAA- ........((((...((((((((((...(((....---.)))..)))))))))).................((((((.........)))))).....----))))....- ( -21.40, z-score = -1.37, R) >droPer1.super_5 3161118 102 - 6813705 ACAACGUAAUUGCACAACGUAAUGGGCAAUCACGC---AGAUGAUCAUUACGUUACACAUACACAAAGAAACACGUGAGACACUGGCACGUGAUGAA----CAAUAGAA- ........((((...((((((((((...(((....---.)))..)))))))))).................((((((.........)))))).....----))))....- ( -21.40, z-score = -1.37, R) >droWil1.scaffold_180708 7319827 97 + 12563649 --------ACGGCACGUA-----AUGUAUUUACGUUUAGUGAGAGCAUUACGUUGCACACACACACAUACAUAUUUUUUUUCUUAAAUAGAAAUGUGUAAUGAGUGUAAA --------...(((((((-----((((.(((((.....))))).)))))))).)))....((((.(((((((((((((..........)))))))))).))).))))... ( -31.20, z-score = -4.66, R) >droVir3.scaffold_12723 5316798 84 + 5802038 ACAACGUUGAUGCGUUUG-----CAGCAAUUACGU---AAGUGUUCAAUACGUUGCACAUACAAAAAUUAA-A--UAAUAAA-AAACACGUUACGG-------------- ....(((.((((.(((((-----((((....((..---..))((.....)))))))...........(((.-.--...))).-)))).))))))).-------------- ( -13.50, z-score = 0.50, R) >droGri2.scaffold_15252 13117338 86 + 17193109 ACAACGUUGUUGCGUUUA-----CGGCAAUUACGU---AAGUGUUCAUUACGUUACACAUACAAAAACAAA-AAUUUAAAAA-AAACACGUAACGG-------------- .......(((((((((((-----((.......)))---))((((..........)))).............-..........-....)))))))).-------------- ( -14.90, z-score = -0.21, R) >consensus ACAACGCAAUUGCAUAGA______GGCAAUCACGC___AGGUGAUCAUUACGUUACACAUACACAAAGAAACACGUGAGACACUUGCACGUGAUGAACGAACGAUAGAA_ ....((..(((((............)))))..))..........(((((((((..................................))))))))).............. ( -7.76 = -8.30 + 0.54)
| Location | 13,487,096 – 13,487,199 |
|---|---|
| Length | 103 |
| Sequences | 12 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 79.70 |
| Shannon entropy | 0.42853 |
| G+C content | 0.42309 |
| Mean single sequence MFE | -29.22 |
| Consensus MFE | -16.64 |
| Energy contribution | -16.57 |
| Covariance contribution | -0.07 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.867931 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
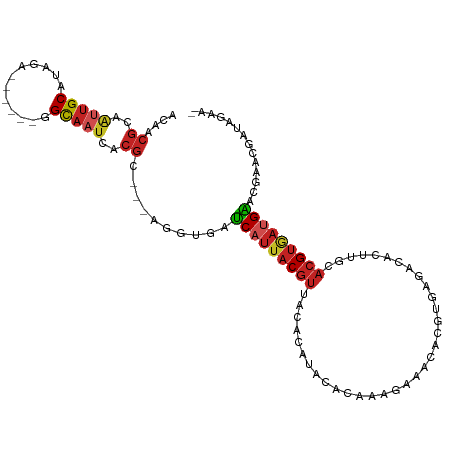
>dm3.chr2L 13487096 103 + 23011544 ACGUGUUUCUUUGUAUAUGUGUAACGUAAUGAUCACCUGCGUGAUUGCC------UCUAUGCAAUU-GCGCUGUGCCGCUUACGUUAC---CGUCUUGCAGUU-GGUUGUAAUU ..........(((((.(((.(((((((((.(..(((..(((..(((((.------.....))))).-.))).)))...))))))))))---)))..)))))..-.......... ( -32.10, z-score = -3.24, R) >droSim1.chr2L 13268979 103 + 22036055 ACGUGUUUCUUUGUAUAUGUGUAACGUAAUGAUCACCUGCGUGAUUGCC------UCUAUGCAAUU-GCGUUGUGCCGCUUACGUUAC---CGUCUUGCAGUU-GGUUGUAAUU ..........(((((.(((.(((((((((.(..(((..(((..(((((.------.....))))).-.))).)))...))))))))))---)))..)))))..-.......... ( -30.40, z-score = -2.78, R) >droSec1.super_16 1632354 103 + 1878335 ACGUGUUUCUUUGUAUAUGUGUAACGUAAUGAUCACCUGCGUGAUUGCC------UCUAUGCAAUU-GCGUUGUGCCGCUUACGUUAC---CGUCUUGCAGUU-GGUUGUAAUU ..........(((((.(((.(((((((((.(..(((..(((..(((((.------.....))))).-.))).)))...))))))))))---)))..)))))..-.......... ( -30.40, z-score = -2.78, R) >droYak2.chr2L 9915270 103 + 22324452 ACGUGUUUCUUUGUAUAUGUGUAACGUAAUGAUCACCUGCGUGAUUGCA------UCUAUGCAAUU-GCGCUGUGCCGCUUACGUUGC---CGUCUUGCAGUU-AGUUGUAAUU ..........(((((.(((.(((((((((.(..(((..(((..((((((------....)))))).-.))).)))...))))))))))---)))..)))))..-.......... ( -33.50, z-score = -3.19, R) >droEre2.scaffold_4929 14678912 103 - 26641161 ACGUGUUUCUUUGUAUAUGUGUAACGUAAUGAUCACCUACGUGAUUGCC------UCUAUGCAAUU-GCGCUGUGCCGCUUACGUUGC---CGUCUUGCAGUU-GGUUGUAAUU ..........(((((.(((.(((((((((.(..(((...((..(((((.------.....))))).-.))..)))...))))))))))---)))..)))))..-.......... ( -27.70, z-score = -1.67, R) >droAna3.scaffold_12943 2191362 103 + 5039921 ACGUGAUUCUUUGUGUAUGUGUAACGUAAUGAUCACCUGCGUGGUUGCC------GCAAUGCAAUG-GCGUAGUGUCGCUUACGUUAC---CGUCUUGCAGUU-GGUUGUAAUU ..(..(((..(((((.(((.((((((((((((.(((.(((((.(((((.------.....))))).-))))))))))).)))))))))---)))..)))))..-)))..).... ( -35.70, z-score = -2.57, R) >dp4.chr4_group1 3212744 109 - 5278887 ACGUGUUUCUUUGUGUAUGUGUAACGUAAUGAUCAUCUGCGUGAUUGCCCAUUACGUUGUGCAAUU-ACGUUGUGCCGCUUACGUUGC---CGUCUUGCAGUU-GGUUGUAAUU ..........(((((.(((.(((((((((.(..(((..((((((((((.((......)).))))))-)))).)))...))))))))))---)))..)))))..-.......... ( -30.10, z-score = -1.48, R) >droPer1.super_5 3161147 109 + 6813705 ACGUGUUUCUUUGUGUAUGUGUAACGUAAUGAUCAUCUGCGUGAUUGCCCAUUACGUUGUGCAAUU-ACGUUGUGCCGCUUACGUUGC---CGUCUUGCAGUU-GGUUGUAAUU ..........(((((.(((.(((((((((.(..(((..((((((((((.((......)).))))))-)))).)))...))))))))))---)))..)))))..-.......... ( -30.10, z-score = -1.48, R) >droWil1.scaffold_180708 7319861 103 - 12563649 AAAUAUGUAUGUGUGUGUGUGCAACGUAAUG--CUCUCAC-UAAACGUA--------AAUACAUUACGUGCCGUGUCGCUUACGUUGCUCCCGUCUUGCAGUUUGGUUGUAAUU ..(((..((....))..)))(((((((((.(--(...(((-...(((((--------(.....))))))...)))..))))))))))).......((((((.....)))))).. ( -26.20, z-score = -1.25, R) >droVir3.scaffold_12723 5316818 99 - 5802038 ----UUUAAUUUUUGUAUGUGCAACGUAUUGAACACUUACGUAAUUGCU-----GCAAACGCAUCA-ACGUUGUGCCAUU-ACGUUGC---CGUCUUGCAGUU-GGUUGUAAUU ----..(((((.(((((((.((((((((.((..(((..((((...(((.-----......)))...-)))).))).)).)-)))))))---)))...))))..-)))))..... ( -25.20, z-score = -0.67, R) >droMoj3.scaffold_6500 3333450 93 - 32352404 -UUUCAAUUUGUAUGUGUGCGUAACGUAAUGGACACUUACGUG---------------ACGCAAUA-GCGUUGUUCCAUUCACGUUGC---CGUCUUGCAGUU-GGUUGUAAUU -..((((((.(((...(((.((((((((((((((((....)))---------------((((....-))))...)))))).)))))))---)).).)))))))-))........ ( -25.90, z-score = -1.09, R) >droGri2.scaffold_15252 13117358 102 - 17193109 --AUUUUUGUUUUUGUAUGUGUAACGUAAUGAACACUUACGUAAUUGCC-----GUAAACGCAACA-ACGUUGUGCCGUUUACGUUGC---CGUCUUGCAGUU-GGUUGUAAUU --..............(((.(((((((((....(((..((((..((((.-----......))))..-)))).)))....)))))))))---))).((((((..-..)))))).. ( -23.30, z-score = 0.19, R) >consensus ACGUGUUUCUUUGUGUAUGUGUAACGUAAUGAUCACCUGCGUGAUUGCC______UCUAUGCAAUU_GCGUUGUGCCGCUUACGUUGC___CGUCUUGCAGUU_GGUUGUAAUU ....................(((((((((....(((..((((..((((............))))...)))).)))....))))))))).......((((((.....)))))).. (-16.64 = -16.57 + -0.07)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:38:02 2011