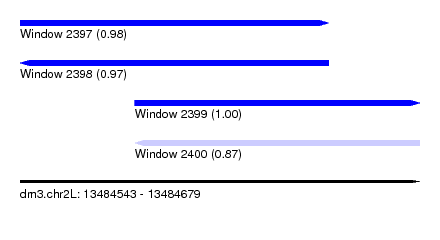
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 13,484,543 – 13,484,679 |
| Length | 136 |
| Max. P | 0.998624 |

| Location | 13,484,543 – 13,484,648 |
|---|---|
| Length | 105 |
| Sequences | 7 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 85.67 |
| Shannon entropy | 0.27165 |
| G+C content | 0.58522 |
| Mean single sequence MFE | -28.16 |
| Consensus MFE | -21.19 |
| Energy contribution | -21.68 |
| Covariance contribution | 0.49 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.19 |
| SVM RNA-class probability | 0.984995 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
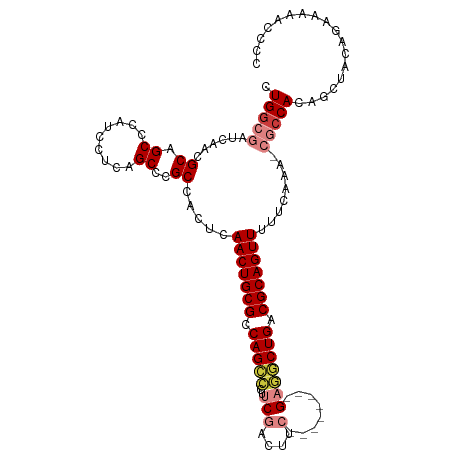
>dm3.chr2L 13484543 105 + 23011544 CUGGCGAUCAACGCAGCCCAUCCUCAGCCCGCCACUCAACUGCGCCAGCCCUUCGACUUC--------GAGGCUGACGCAGUUUUUCAAA-CGCCACAGCUACAGAAAAACCCC .(((((......((.((.........))..)).....(((((((.(((((..(((....)--------))))))).))))))).......-))))).................. ( -30.60, z-score = -3.19, R) >droSim1.chr2L 13266422 105 + 22036055 CUGGCGAUCAACGCAGCCCAUCCUCAGCCCGCCACUCAACUGCGCCAGCCCUUCGACUUC--------GAGGCUGACGCAGUUUUUCAAA-CGCCACAGCUACAGAAAAACCCC .(((((......((.((.........))..)).....(((((((.(((((..(((....)--------))))))).))))))).......-))))).................. ( -30.60, z-score = -3.19, R) >droSec1.super_16 1629816 104 + 1878335 CUGGCGAUCAACGCAGCCCAUCCUCAGCCCGCCACUCAACUGCGCCAGCCCUUCGACUUC--------GAGGCUGACGCAGUUUUUCAAA-CGCCACAGCUACAGAAAAACCC- .(((((......((.((.........))..)).....(((((((.(((((..(((....)--------))))))).))))))).......-))))).................- ( -30.60, z-score = -3.11, R) >droYak2.chr2L 9912689 105 + 22324452 CUGGCGAUCAACGCAGCCCAUCCUCAGCCCGCCACUCAACUGCGCCAGCCCUUCGACUCC--------GAGGCUGACGCAGUUUUUCAAA-CGCCACAGCUACAGAAAAACCCC .(((((......((.((.........))..)).....(((((((.(((((..(((....)--------))))))).))))))).......-))))).................. ( -30.00, z-score = -3.05, R) >droEre2.scaffold_4929 14676358 105 - 26641161 CUGGCGAUCAACGCAGCCCAUCCUCAGCCCGCCACUCAACUGCGCCAGCCCUUCGAUUCC--------GAGGCUGACGCAGUUUUUCAAA-CGCCACAGCUACAGAAAAACCCC .(((((......((.((.........))..)).....(((((((.(((((..(((....)--------))))))).))))))).......-))))).................. ( -30.00, z-score = -3.12, R) >droAna3.scaffold_12943 2188770 95 + 5039921 CUGGCGAUCAACGCAGCCCAUCCUCAGCCCGCCACUCAACUGCGCCAG----UCACUUCC--------G--GCUGACGCAGUUUUCCAAC-AACCA-GGCUACAGAAAGAA--- ((((((.....)))((((........(....).....(((((((.(((----((......--------)--)))).))))))).......-.....-)))).)))......--- ( -25.90, z-score = -1.67, R) >droWil1.scaffold_180708 7317033 109 - 12563649 CUGUCGAUCAACGCAGCCCAUCCUCAGCCCGCCACUCAACUGCGCCAGUGCCCCUCUCUCUCUACUCAGUCACUGACGCAGUUUCUCAACCAAUCACAGCAGCAAACAA----- ((((.(((....((.((.........))..)).....(((((((.(((((.(................).))))).))))))).........)))))))..........----- ( -19.39, z-score = -1.48, R) >consensus CUGGCGAUCAACGCAGCCCAUCCUCAGCCCGCCACUCAACUGCGCCAGCCCUUCGACUUC________GAGGCUGACGCAGUUUUUCAAA_CGCCACAGCUACAGAAAAACCCC .(((((......((.((.........))..)).....(((((((.(((((....................))))).)))))))........))))).................. (-21.19 = -21.68 + 0.49)

| Location | 13,484,543 – 13,484,648 |
|---|---|
| Length | 105 |
| Sequences | 7 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 85.67 |
| Shannon entropy | 0.27165 |
| G+C content | 0.58522 |
| Mean single sequence MFE | -42.57 |
| Consensus MFE | -34.91 |
| Energy contribution | -34.80 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.88 |
| SVM RNA-class probability | 0.972789 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
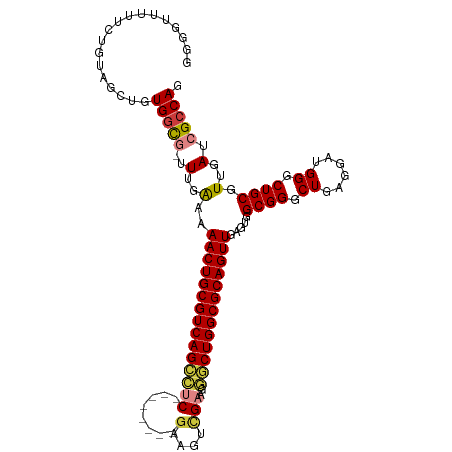
>dm3.chr2L 13484543 105 - 23011544 GGGGUUUUUCUGUAGCUGUGGCG-UUUGAAAAACUGCGUCAGCCUC--------GAAGUCGAAGGGCUGGCGCAGUUGAGUGGCGGGCUGAGGAUGGGCUGCGUUGAUCGCCAG ..((((.......)))).(((((-.(..(..(((((((((((((((--------(....)))..))))))))))))).....((((.((......)).)))).)..).))))). ( -44.80, z-score = -2.50, R) >droSim1.chr2L 13266422 105 - 22036055 GGGGUUUUUCUGUAGCUGUGGCG-UUUGAAAAACUGCGUCAGCCUC--------GAAGUCGAAGGGCUGGCGCAGUUGAGUGGCGGGCUGAGGAUGGGCUGCGUUGAUCGCCAG ..((((.......)))).(((((-.(..(..(((((((((((((((--------(....)))..))))))))))))).....((((.((......)).)))).)..).))))). ( -44.80, z-score = -2.50, R) >droSec1.super_16 1629816 104 - 1878335 -GGGUUUUUCUGUAGCUGUGGCG-UUUGAAAAACUGCGUCAGCCUC--------GAAGUCGAAGGGCUGGCGCAGUUGAGUGGCGGGCUGAGGAUGGGCUGCGUUGAUCGCCAG -.((((.......)))).(((((-.(..(..(((((((((((((((--------(....)))..))))))))))))).....((((.((......)).)))).)..).))))). ( -44.80, z-score = -2.60, R) >droYak2.chr2L 9912689 105 - 22324452 GGGGUUUUUCUGUAGCUGUGGCG-UUUGAAAAACUGCGUCAGCCUC--------GGAGUCGAAGGGCUGGCGCAGUUGAGUGGCGGGCUGAGGAUGGGCUGCGUUGAUCGCCAG ..((((.......)))).(((((-.(..(..(((((((((((((((--------(....)))..))))))))))))).....((((.((......)).)))).)..).))))). ( -43.90, z-score = -2.07, R) >droEre2.scaffold_4929 14676358 105 + 26641161 GGGGUUUUUCUGUAGCUGUGGCG-UUUGAAAAACUGCGUCAGCCUC--------GGAAUCGAAGGGCUGGCGCAGUUGAGUGGCGGGCUGAGGAUGGGCUGCGUUGAUCGCCAG ..((((.......)))).(((((-.(..(..(((((((((((((((--------(....)))..))))))))))))).....((((.((......)).)))).)..).))))). ( -43.90, z-score = -2.32, R) >droAna3.scaffold_12943 2188770 95 - 5039921 ---UUCUUUCUGUAGCC-UGGUU-GUUGGAAAACUGCGUCAGC--C--------GGAAGUGA----CUGGCGCAGUUGAGUGGCGGGCUGAGGAUGGGCUGCGUUGAUCGCCAG ---.(((.(((.(((((-((.(.-.((....((((((((((((--(--------....).).----))))))))))).))..)))))))).))).)))..(((.....)))... ( -38.90, z-score = -2.03, R) >droWil1.scaffold_180708 7317033 109 + 12563649 -----UUGUUUGCUGCUGUGAUUGGUUGAGAAACUGCGUCAGUGACUGAGUAGAGAGAGAGGGGCACUGGCGCAGUUGAGUGGCGGGCUGAGGAUGGGCUGCGUUGAUCGACAG -----..........(((((((..(......(((((((((((((.((.............))..))))))))))))).....((((.((......)).)))).)..))).)))) ( -36.92, z-score = -1.67, R) >consensus GGGGUUUUUCUGUAGCUGUGGCG_UUUGAAAAACUGCGUCAGCCUC________GAAGUCGAAGGGCUGGCGCAGUUGAGUGGCGGGCUGAGGAUGGGCUGCGUUGAUCGCCAG ..................((((.........((((((((((((((..................)))))))))))))).....((((.((......)).)))).......)))). (-34.91 = -34.80 + -0.11)

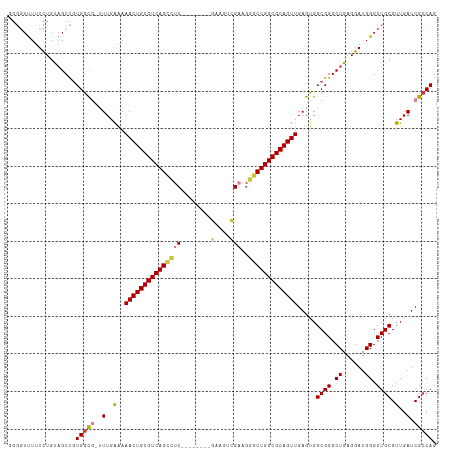
| Location | 13,484,582 – 13,484,679 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 98.56 |
| Shannon entropy | 0.02489 |
| G+C content | 0.52685 |
| Mean single sequence MFE | -22.26 |
| Consensus MFE | -22.26 |
| Energy contribution | -22.26 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -4.00 |
| Structure conservation index | 1.00 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.42 |
| SVM RNA-class probability | 0.998624 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2L 13484582 97 + 23011544 CUGCGCCAGCCCUUCGACUUCGAGGCUGACGCAGUUUUUCAAACGCCACAGCUACAGAAAAACCCCCCAAAAAACCAACCACCACAAGAACUACAGC .((((.(((((..(((....)))))))).))))(((((((....((....))....))))))).................................. ( -22.50, z-score = -4.07, R) >droSim1.chr2L 13266461 97 + 22036055 CUGCGCCAGCCCUUCGACUUCGAGGCUGACGCAGUUUUUCAAACGCCACAGCUACAGAAAAACCCCCCAAAAAACCAACCACCACAAGAACUACAGC .((((.(((((..(((....)))))))).))))(((((((....((....))....))))))).................................. ( -22.50, z-score = -4.07, R) >droSec1.super_16 1629855 96 + 1878335 CUGCGCCAGCCCUUCGACUUCGAGGCUGACGCAGUUUUUCAAACGCCACAGCUACAGAAAAACCCCC-AAAAAACCAACCACCACAAGAACUACAGC .((((.(((((..(((....)))))))).))))(((((((....((....))....)))))))....-............................. ( -22.50, z-score = -3.80, R) >droYak2.chr2L 9912728 97 + 22324452 CUGCGCCAGCCCUUCGACUCCGAGGCUGACGCAGUUUUUCAAACGCCACAGCUACAGAAAAACCCCCCAAAAAACCAACCACCACAAGAACUACAGC .((((.(((((..(((....)))))))).))))(((((((....((....))....))))))).................................. ( -21.90, z-score = -3.97, R) >droEre2.scaffold_4929 14676397 97 - 26641161 CUGCGCCAGCCCUUCGAUUCCGAGGCUGACGCAGUUUUUCAAACGCCACAGCUACAGAAAAACCCCCCAAAAAACCAACCACCACAAGAACUACAGC .((((.(((((..(((....)))))))).))))(((((((....((....))....))))))).................................. ( -21.90, z-score = -4.09, R) >consensus CUGCGCCAGCCCUUCGACUUCGAGGCUGACGCAGUUUUUCAAACGCCACAGCUACAGAAAAACCCCCCAAAAAACCAACCACCACAAGAACUACAGC .((((.(((((..(((....)))))))).))))(((((((....((....))....))))))).................................. (-22.26 = -22.26 + -0.00)

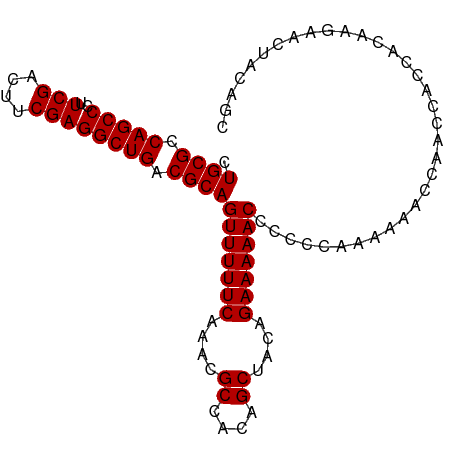
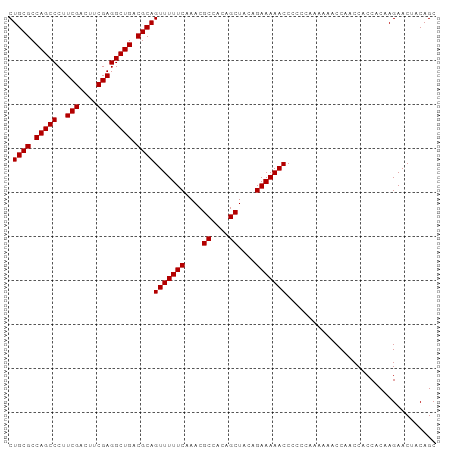
| Location | 13,484,582 – 13,484,679 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 98.56 |
| Shannon entropy | 0.02489 |
| G+C content | 0.52685 |
| Mean single sequence MFE | -31.26 |
| Consensus MFE | -31.24 |
| Energy contribution | -31.24 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.37 |
| Structure conservation index | 1.00 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.866260 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2L 13484582 97 - 23011544 GCUGUAGUUCUUGUGGUGGUUGGUUUUUUGGGGGGUUUUUCUGUAGCUGUGGCGUUUGAAAAACUGCGUCAGCCUCGAAGUCGAAGGGCUGGCGCAG ((..(((...)))..))................((((((((....((....))....))))))))((((((((((((....)))..))))))))).. ( -31.60, z-score = -1.49, R) >droSim1.chr2L 13266461 97 - 22036055 GCUGUAGUUCUUGUGGUGGUUGGUUUUUUGGGGGGUUUUUCUGUAGCUGUGGCGUUUGAAAAACUGCGUCAGCCUCGAAGUCGAAGGGCUGGCGCAG ((..(((...)))..))................((((((((....((....))....))))))))((((((((((((....)))..))))))))).. ( -31.60, z-score = -1.49, R) >droSec1.super_16 1629855 96 - 1878335 GCUGUAGUUCUUGUGGUGGUUGGUUUUUU-GGGGGUUUUUCUGUAGCUGUGGCGUUUGAAAAACUGCGUCAGCCUCGAAGUCGAAGGGCUGGCGCAG ((..((((((....))............(-(.(((....))).))))))..))..........((((((((((((((....)))..))))))))))) ( -31.70, z-score = -1.61, R) >droYak2.chr2L 9912728 97 - 22324452 GCUGUAGUUCUUGUGGUGGUUGGUUUUUUGGGGGGUUUUUCUGUAGCUGUGGCGUUUGAAAAACUGCGUCAGCCUCGGAGUCGAAGGGCUGGCGCAG ((..(((...)))..))................((((((((....((....))....))))))))((((((((((((....)))..))))))))).. ( -30.70, z-score = -1.04, R) >droEre2.scaffold_4929 14676397 97 + 26641161 GCUGUAGUUCUUGUGGUGGUUGGUUUUUUGGGGGGUUUUUCUGUAGCUGUGGCGUUUGAAAAACUGCGUCAGCCUCGGAAUCGAAGGGCUGGCGCAG ((..(((...)))..))................((((((((....((....))....))))))))((((((((((((....)))..))))))))).. ( -30.70, z-score = -1.23, R) >consensus GCUGUAGUUCUUGUGGUGGUUGGUUUUUUGGGGGGUUUUUCUGUAGCUGUGGCGUUUGAAAAACUGCGUCAGCCUCGAAGUCGAAGGGCUGGCGCAG ((..(((...)))..))................((((((((....((....))....))))))))((((((((((((....)))..))))))))).. (-31.24 = -31.24 + 0.00)

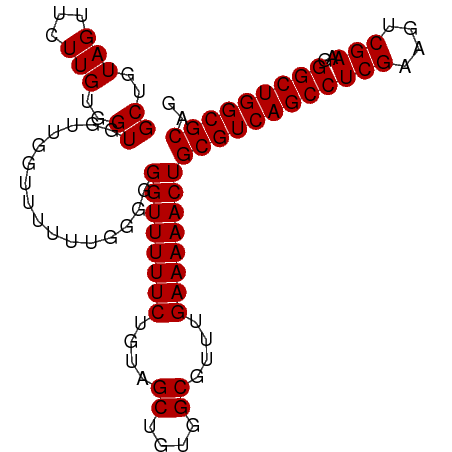
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:38:01 2011