
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 13,465,375 – 13,465,487 |
| Length | 112 |
| Max. P | 0.749536 |

| Location | 13,465,375 – 13,465,483 |
|---|---|
| Length | 108 |
| Sequences | 7 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 62.94 |
| Shannon entropy | 0.74223 |
| G+C content | 0.36137 |
| Mean single sequence MFE | -25.37 |
| Consensus MFE | -7.48 |
| Energy contribution | -9.03 |
| Covariance contribution | 1.55 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.29 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.749536 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 13465375 108 - 23011544 CCUGGUCUACAGAUAUUGCAUUUGUUGCAAUGCAAAUGCAGCGAGGCUUUUAAGACCGAACUAUAAAUUGGAUUUCUUUUUACCGGAAGCGG---AAAUGUUUGUAGAAAU .....((((((((((((((.(((((((((.......)))))))))))........((((........))))...........(((....)))---.))))))))))))... ( -30.60, z-score = -2.79, R) >droAna3.scaffold_12943 2169599 92 - 5039921 ------------------CAGGCUUCA-GGAGCGAGAGCAAAUCGAGUGUGAGUGCGGCUGUAAAUUUUAUUUUUUUCUUUCCAAGAACUUGUUCAAGUCAUGGUAAGAUU ------------------(((((((((-.(..(((.......)))..).)))).))..)))...............((((.(((.((.(((....))))).))).)))).. ( -18.00, z-score = 0.76, R) >droEre2.scaffold_4929 14657833 108 + 26641161 CCUGGUCUGCAGAUACUUUAUUUGUUGCAGUGCAAAUGCAGCGCGGCUUUUAAGACCUAACUAUAAAUUGGAUUUCUAUUUACCAAAAGCGG---AAAUCUUUGUAGAAAU ...((((((((((((...))))))).((.((((.......)))).)).....)))))...((((((...(((((((..(((.....)))..)---)))))))))))).... ( -27.90, z-score = -2.00, R) >droYak2.chr2L 9893555 108 - 22324452 CCUGGUCUACAGAUAUUGUGUUUGUUGCAAUGCAAAUGCAGCGAGGCUUUUAAGACCAAACUAUAAAUUGCAUUUCUAUUUACCAGAAGUGG---AAAUAUUUGUAGGAAU ..((((((((((...)))).(((((((((.......))))))))).......))))))..((((((((...((((((((((.....))))))---)))))))))))).... ( -29.00, z-score = -2.45, R) >droSec1.super_16 1611420 108 - 1878335 CCUGGUCUACAGAUAUUGCAUUUGUUGCAAUGCAAAUGCAGAGAGGCUUUUAAGACCGAACUAUAAAUUGGAUUUCUAUUUAGCAGAAGCGG---AAAUGUGUGUAGAAAU .....((((((.(((((((((((((......))))))))................(((..((.(((((.((....)))))))..))...)))---.))))).))))))... ( -27.50, z-score = -1.86, R) >droSim1.chr2L 13247682 106 - 22036055 CCUGGUCUACAGAUAUUGCAUUUGUUGCAAUGCAAAUGCAGUGAGGCUUUUAAGACCGAACUAUAAAUUGGAUUUCUAUUC--CAGAAACGG---AAAUGUUUGUAGAAAU .....((((((((((((((((((((......))))))))(((..((((....)).))..))).....(((((.......))--)))......---.))))))))))))... ( -31.20, z-score = -3.67, R) >droMoj3.scaffold_6500 3304042 91 + 32352404 ------CAGCACUUUCAGCAGUUUUCACUUGGAAAUAACUUUAUGAGUUUGAAG--CUAGUUAAAUAAUUGAUUUCAUUUAACUAAUUUUGA---UGGCCUU--------- ------..((((((..((..((((((....))))))..))....))))(..(((--.(((((((((.(......).))))))))).)))..)---..))...--------- ( -13.40, z-score = 0.41, R) >consensus CCUGGUCUACAGAUAUUGCAUUUGUUGCAAUGCAAAUGCAGCGAGGCUUUUAAGACCGAACUAUAAAUUGGAUUUCUAUUUACCAGAAGCGG___AAAUCUUUGUAGAAAU ...(((((..((...((((((((((......)))))))))).....))....)))))...................................................... ( -7.48 = -9.03 + 1.55)
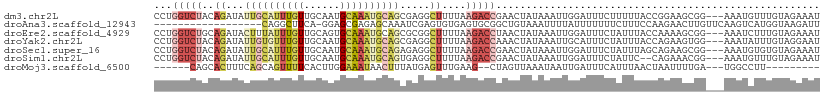

| Location | 13,465,379 – 13,465,487 |
|---|---|
| Length | 108 |
| Sequences | 7 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 61.10 |
| Shannon entropy | 0.78813 |
| G+C content | 0.36930 |
| Mean single sequence MFE | -23.60 |
| Consensus MFE | -7.90 |
| Energy contribution | -9.51 |
| Covariance contribution | 1.62 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.04 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.539458 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 13465379 108 - 23011544 CAGACCUGGUCUACAGAUAUUGCAUUUGUUGCAAUGCAAAUGCAGCGAGGCUUUUAAGACCGAACUAUAAAUUGGAUUUCUUUUUACCGGAAGCGGAAAUGUUUGUAG--- ..........(((((((((((((.(((((((((.......)))))))))))........((((........))))...........(((....))).)))))))))))--- ( -27.80, z-score = -1.76, R) >droAna3.scaffold_12943 2169603 92 - 5039921 ------------------CAGGCAGGCUUCA-GGAGCGAGAGCAAAUCGAGUGUGAGUGCGGCUGUAAAUUUUAUUUUUUUCUUUCCAAGAACUUGUUCAAGUCAUGGUAA ------------------..(((..((((((-.(..(((.......)))..).)))).)).))).....................(((.((.(((....))))).)))... ( -17.30, z-score = 1.38, R) >droEre2.scaffold_4929 14657837 108 + 26641161 CAAACCUGGUCUGCAGAUACUUUAUUUGUUGCAGUGCAAAUGCAGCGCGGCUUUUAAGACCUAACUAUAAAUUGGAUUUCUAUUUACCAAAAGCGGAAAUCUUUGUAG--- .......((((((((((((...))))))).((.((((.......)))).)).....)))))...((((((...(((((((..(((.....)))..)))))))))))))--- ( -26.80, z-score = -1.77, R) >droYak2.chr2L 9893559 108 - 22324452 CAAGCCUGGUCUACAGAUAUUGUGUUUGUUGCAAUGCAAAUGCAGCGAGGCUUUUAAGACCAAACUAUAAAUUGCAUUUCUAUUUACCAGAAGUGGAAAUAUUUGUAG--- .((((((.((..........(((((((((......))))))))))).))))))...........((((((((...((((((((((.....))))))))))))))))))--- ( -28.10, z-score = -2.02, R) >droSec1.super_16 1611424 108 - 1878335 CAAACCUGGUCUACAGAUAUUGCAUUUGUUGCAAUGCAAAUGCAGAGAGGCUUUUAAGACCGAACUAUAAAUUGGAUUUCUAUUUAGCAGAAGCGGAAAUGUGUGUAG--- ..........(((((.(((((((((((((......))))))))................(((..((.(((((.((....)))))))..))...))).))))).)))))--- ( -24.70, z-score = -0.82, R) >droSim1.chr2L 13247686 106 - 22036055 CAAACCUGGUCUACAGAUAUUGCAUUUGUUGCAAUGCAAAUGCAGUGAGGCUUUUAAGACCGAACUAUAAAUUGGAUUUCUAUUC--CAGAAACGGAAAUGUUUGUAG--- ......((((((..((.((((((((((((......))))))))))))...))....))))))..(((((((((((....)))(((--(......))))..))))))))--- ( -28.60, z-score = -2.79, R) >droMoj3.scaffold_6500 3304046 91 + 32352404 ------AAAACAGCACUUUCAGCAGUUUUCACUUGGAAAUAACUUUAUGAGUUUGAAG--CUAGUUAAAUAAUUGAUUUCAUUUAACUAAUUUUGAUGG------------ ------........((((..((..((((((....))))))..))....))))(..(((--.(((((((((.(......).))))))))).)))..)...------------ ( -11.90, z-score = 0.52, R) >consensus CAAACCUGGUCUACAGAUAUUGCAUUUGUUGCAAUGCAAAUGCAGCGAGGCUUUUAAGACCGAACUAUAAAUUGGAUUUCUAUUUACCAGAAGCGGAAAUGUUUGUAG___ .......(((((..((...((((((((((......)))))))))).....))....))))).................................................. ( -7.90 = -9.51 + 1.62)
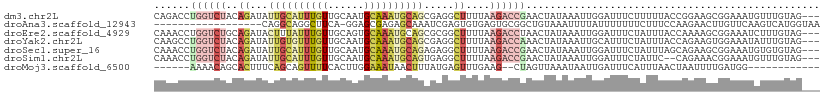

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:37:54 2011