
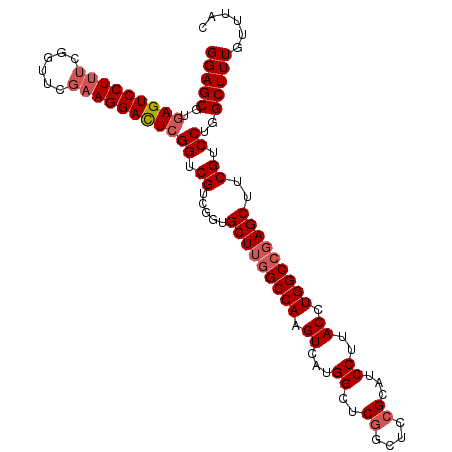
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 13,461,389 – 13,461,496 |
| Length | 107 |
| Max. P | 0.970727 |

| Location | 13,461,389 – 13,461,496 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 92.70 |
| Shannon entropy | 0.12843 |
| G+C content | 0.60607 |
| Mean single sequence MFE | -44.02 |
| Consensus MFE | -37.02 |
| Energy contribution | -38.06 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.658876 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 13461389 107 + 23011544 GGAGCGUGAGUCCUUUCGGUUCGAAGGACUCGGUCGUCGGUGCUUGGCCAAGUCAUGGCUCGGCUCCGCAUCCUUACCUGGCCGAGCUUCGUCCUGGCUUUGUUUAC (((((..(((((((((......)))))))))((....(((.(((((((((.((...((..((....))...))..)).))))))))).))).))..)))))...... ( -43.20, z-score = -1.88, R) >droSim1.chr2L 13243713 107 + 22036055 GGAGCGUGAGUCCUUUCGGUUCGAAGGAUUCGGUCGUCGGUGCUUGGCCAAGUCAUGGCUCGGCUCGGCAUCCUUACCUGGCCGAGCUUCGUCCUGGCUUUGUUUAC .(((((.(((((((((......)))))))))(((((........)))))((((((.(((..((((((((...........))))))))..))).))))))))))).. ( -42.40, z-score = -1.70, R) >droSec1.super_16 1607461 107 + 1878335 GGAGCGUGAGUCCUUUCGGUUCGAAGGACUCGGGCGUCGGUGCUUGGCCAAGUCAUGGCUCGGCUUCGCAACCUUACCUGGCCGAGCUUCGUCCUGGCUUUGUUUAC (..((.((((((((((......)))))))))).))..)...((..(((((.(.(..(((((((((..(........)..)))))))))..).).)))))..)).... ( -46.50, z-score = -2.47, R) >droYak2.chr2L 9889251 106 + 22324452 GGAGCGGAAGUCCUUC-GGUUCUCAGGACUCGGGCGUCUCUGCUUGGCCAAGUCAUGGCUCGGCUCCGCAUCCUUACCUGGCCGAGCUUCGUCCUGGCUUUGUUUAC .(((((((.((((...-((((.....)))).))))...)))))))(((((.(.(..(((((((((..(........)..)))))))))..).).)))))........ ( -40.20, z-score = -1.11, R) >droEre2.scaffold_4929 14653840 106 - 26641161 GGAGCGGGAGUCCUUU-GGCUCGAAGGACUCGGUCGUCGGUGCUGGGCCAAGUCACGGCUCGGCUCCGCAUCCCUACCUGGCGGAGCUUCGUCCUGGCUUUGUUUAC ((((.(((((((((((-(...))))))))))....(.(((.((((((((.......)))))))).))))..)))).)).(((((((((.......)))))))))... ( -47.80, z-score = -1.61, R) >consensus GGAGCGUGAGUCCUUUCGGUUCGAAGGACUCGGUCGUCGGUGCUUGGCCAAGUCAUGGCUCGGCUCCGCAUCCUUACCUGGCCGAGCUUCGUCCUGGCUUUGUUUAC (((((..(((((((((......)))))))))((.((.....(((((((((.((...((..((....))...))..)).)))))))))..)).))..)))))...... (-37.02 = -38.06 + 1.04)


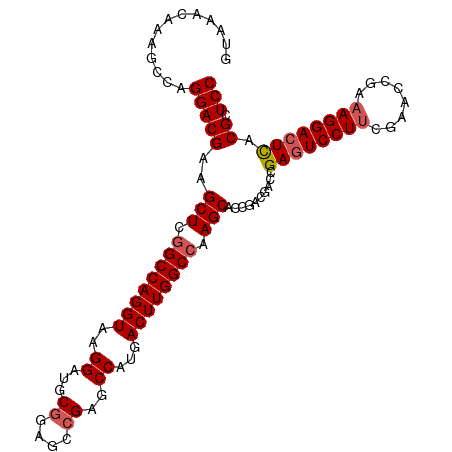
| Location | 13,461,389 – 13,461,496 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 92.70 |
| Shannon entropy | 0.12843 |
| G+C content | 0.60607 |
| Mean single sequence MFE | -38.74 |
| Consensus MFE | -35.04 |
| Energy contribution | -35.68 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.90 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.84 |
| SVM RNA-class probability | 0.970727 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 13461389 107 - 23011544 GUAAACAAAGCCAGGACGAAGCUCGGCCAGGUAAGGAUGCGGAGCCGAGCCAUGACUUGGCCAAGCACCGACGACCGAGUCCUUCGAACCGAAAGGACUCACGCUCC .............(((((..(((.((((((((..((...((....))..))...)))))))).)))..........((((((((((...)).)))))))).)).))) ( -40.70, z-score = -3.05, R) >droSim1.chr2L 13243713 107 - 22036055 GUAAACAAAGCCAGGACGAAGCUCGGCCAGGUAAGGAUGCCGAGCCGAGCCAUGACUUGGCCAAGCACCGACGACCGAAUCCUUCGAACCGAAAGGACUCACGCUCC .........((((((.((..(((((((..((((....))))..)))))))..)).))))))..(((..(((.(........).)))..((....))......))).. ( -37.80, z-score = -3.03, R) >droSec1.super_16 1607461 107 - 1878335 GUAAACAAAGCCAGGACGAAGCUCGGCCAGGUAAGGUUGCGAAGCCGAGCCAUGACUUGGCCAAGCACCGACGCCCGAGUCCUUCGAACCGAAAGGACUCACGCUCC ........(((..((.((..(((.((((((((..((((.((....))))))...)))))))).)))..))...)).((((((((((...)).))))))))..))).. ( -41.40, z-score = -3.16, R) >droYak2.chr2L 9889251 106 - 22324452 GUAAACAAAGCCAGGACGAAGCUCGGCCAGGUAAGGAUGCGGAGCCGAGCCAUGACUUGGCCAAGCAGAGACGCCCGAGUCCUGAGAACC-GAAGGACUUCCGCUCC .........((((((.((..((((((((..(((....))).).)))))))..)).))))))......(((.((...(((((((.......-..))))))).))))). ( -37.60, z-score = -1.86, R) >droEre2.scaffold_4929 14653840 106 + 26641161 GUAAACAAAGCCAGGACGAAGCUCCGCCAGGUAGGGAUGCGGAGCCGAGCCGUGACUUGGCCCAGCACCGACGACCGAGUCCUUCGAGCC-AAAGGACUCCCGCUCC ........(((..((.((......)))).(((..((.(((((.((((((......)))))))).)))))....)))((((((((......-.))))))))..))).. ( -36.20, z-score = -0.67, R) >consensus GUAAACAAAGCCAGGACGAAGCUCGGCCAGGUAAGGAUGCGGAGCCGAGCCAUGACUUGGCCAAGCACCGACGACCGAGUCCUUCGAACCGAAAGGACUCACGCUCC .............(((((..(((.((((((((..((...((....))..))...)))))))).)))..........((((((((........)))))))).)).))) (-35.04 = -35.68 + 0.64)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:37:52 2011