| Sequence ID | dm3.chr2L |
|---|---|
| Location | 1,250,003 – 1,250,110 |
| Length | 107 |
| Max. P | 0.993225 |

| Location | 1,250,003 – 1,250,110 |
|---|---|
| Length | 107 |
| Sequences | 9 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.53 |
| Shannon entropy | 0.40551 |
| G+C content | 0.34338 |
| Mean single sequence MFE | -14.93 |
| Consensus MFE | -11.02 |
| Energy contribution | -11.01 |
| Covariance contribution | -0.01 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.92 |
| SVM RNA-class probability | 0.975224 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
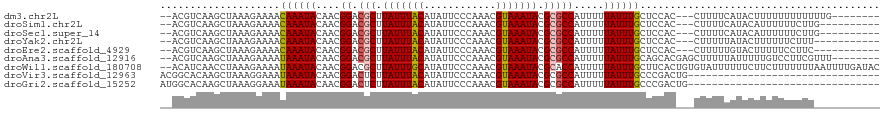
>dm3.chr2L 1250003 107 + 23011544 --ACGUCAAGCUAAAGAAAACAAAUACAACGGACGCUUAUUUACAUAUUCCCAAACGUAAAUACGCGCCAUUUUUAUUUGCUCCAC---CUUUUCAUACUUUUUUUUUUUUG-------- --.............(((((((((((....((.(((.(((((((............))))))).))))).....))))))......---.))))).................-------- ( -14.71, z-score = -2.43, R) >droSim1.chr2L 1222176 105 + 22036055 --ACGUCAAGCUAAAGAAAACAAAUACAACGGACGCUUAUUUACAUAUUCCCAAACGUAAAUACGCGCCAUUUUUAUUUGCUCCAC---CUUUUCAUACAUUUUUUCUUG---------- --...........(((((((((((((....((.(((.(((((((............))))))).))))).....))))))......---.............))))))).---------- ( -15.30, z-score = -2.85, R) >droSec1.super_14 1200775 105 + 2068291 --ACGUCAAGCUAAAGAAAACAAAUACAACGGACGCUUAUUUACAUAUUCCCAAACGUAAAUACGCGCCAUUUUUAUUUGCUCCAC---CUUUUCAUACAUUUUUUCUUG---------- --...........(((((((((((((....((.(((.(((((((............))))))).))))).....))))))......---.............))))))).---------- ( -15.30, z-score = -2.85, R) >droYak2.chr2L 1225096 104 + 22324452 --ACGUCAAGCUAAAGAAAACAAAUACAACGGACGCUUAUUUACAUAUUCCCAAACGUAAAUACGCGCCAUUUUUAUUUGCUCCAC---CUUUUUAUACUUUUUUCUUU----------- --..........((((((((((((((....((.(((.(((((((............))))))).))))).....))))))......---............))))))))----------- ( -15.46, z-score = -2.91, R) >droEre2.scaffold_4929 1291457 104 + 26641161 --ACGUCAAGCUAAAGAAAACAAAUACAACGGACGCUUAUUUACAUAUUCCCAAACGUAAAUACGCGCCAUUUUUAUUUGCUCCAC---CUUUUUGUACUUUUUCCUUC----------- --..((((((..........((((((....((.(((.(((((((............))))))).))))).....))))))......---...)))).))..........----------- ( -14.31, z-score = -1.83, R) >droAna3.scaffold_12916 4711880 110 + 16180835 --ACGUCAAGCUAAAGAAAAUAAAUACAACGGACGCUUAUUUACAUAUUCCCAAACGUAAAUACGCGCCAUUUUUAUUUGCAGCACGAGCUUUUUAUUUUUGUCCUUCGUUU-------- --(((..(((.....(((((((((......((.(((.(((((((............))))))).)))))............(((....))).)))))))))...))))))..-------- ( -18.30, z-score = -1.08, R) >droWil1.scaffold_180708 2863127 118 + 12563649 --ACAUCAACCUAAAGAAAAUAAAUACAACGGACGCUUAUUUGCAUAUUCCCAAACGUAAAUACGCACCAUUUUUAUUUGCUUCACUGUGUAUUUUUUCUUCUUUUUUUAAUUUUGAUAC --..(((((...((((((...(((((((.(((..((.(((((((............))))))).))..((........)).....))))))))))....))))))........))))).. ( -17.50, z-score = -2.54, R) >droVir3.scaffold_12963 7172285 88 + 20206255 ACGGCACAAGCUAAAGGAAAUAAAUACAACGGACUCUUAUUUACAUAUUCCCAAACGUAAAUACGCGCCAUUUUUAUUUGCCCGACUG-------------------------------- ..(((....)))...(((((((((......((.(.(.(((((((............))))))).).)))...))))))).))......-------------------------------- ( -11.20, z-score = -0.32, R) >droGri2.scaffold_15252 10319221 88 + 17193109 AUGGCACAAGCUAAAGGAAAUAAAUACAACGGACUCUUAUUUACAUAUUCCCAAACGUAAAUACGCGCCAUUUUUAUUUGCCCGACUG-------------------------------- .((((....))))..(((((((((......((.(.(.(((((((............))))))).).)))...))))))).))......-------------------------------- ( -12.30, z-score = -0.70, R) >consensus __ACGUCAAGCUAAAGAAAACAAAUACAACGGACGCUUAUUUACAUAUUCCCAAACGUAAAUACGCGCCAUUUUUAUUUGCUCCAC___CUUUUUAUACUUUUUUUUUU___________ ....................((((((....((.(((.(((((((............))))))).))))).....))))))........................................ (-11.02 = -11.01 + -0.01)

| Location | 1,250,003 – 1,250,110 |
|---|---|
| Length | 107 |
| Sequences | 9 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.53 |
| Shannon entropy | 0.40551 |
| G+C content | 0.34338 |
| Mean single sequence MFE | -24.95 |
| Consensus MFE | -20.11 |
| Energy contribution | -20.31 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.60 |
| SVM RNA-class probability | 0.993225 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
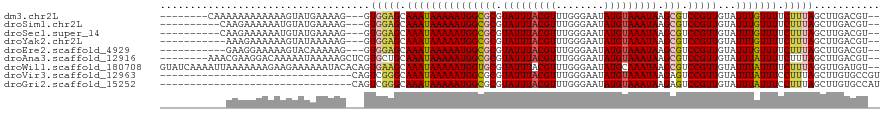
>dm3.chr2L 1250003 107 - 23011544 --------CAAAAAAAAAAAAGUAUGAAAAG---GUGGAGCAAAUAAAAAUGGCGCGUAUUUACGUUUGGGAAUAUGUAAAUAAGCGUCCGUUGUAUUUGUUUUCUUUAGCUUGACGU-- --------.............((..(..(((---(.((((((((((..((((((((.(((((((((........))))))))).))).))))).))))))))))))))..)...))..-- ( -28.50, z-score = -3.72, R) >droSim1.chr2L 1222176 105 - 22036055 ----------CAAGAAAAAAUGUAUGAAAAG---GUGGAGCAAAUAAAAAUGGCGCGUAUUUACGUUUGGGAAUAUGUAAAUAAGCGUCCGUUGUAUUUGUUUUCUUUAGCUUGACGU-- ----------.........((((..(..(((---(.((((((((((..((((((((.(((((((((........))))))))).))).))))).))))))))))))))..)...))))-- ( -29.20, z-score = -3.70, R) >droSec1.super_14 1200775 105 - 2068291 ----------CAAGAAAAAAUGUAUGAAAAG---GUGGAGCAAAUAAAAAUGGCGCGUAUUUACGUUUGGGAAUAUGUAAAUAAGCGUCCGUUGUAUUUGUUUUCUUUAGCUUGACGU-- ----------.........((((..(..(((---(.((((((((((..((((((((.(((((((((........))))))))).))).))))).))))))))))))))..)...))))-- ( -29.20, z-score = -3.70, R) >droYak2.chr2L 1225096 104 - 22324452 -----------AAAGAAAAAAGUAUAAAAAG---GUGGAGCAAAUAAAAAUGGCGCGUAUUUACGUUUGGGAAUAUGUAAAUAAGCGUCCGUUGUAUUUGUUUUCUUUAGCUUGACGU-- -----------..........((.....(((---.(((((.(((((((((((((((.(((((((((........))))))))).))).)))))...))))))).))))).))).))..-- ( -27.50, z-score = -3.41, R) >droEre2.scaffold_4929 1291457 104 - 26641161 -----------GAAGGAAAAAGUACAAAAAG---GUGGAGCAAAUAAAAAUGGCGCGUAUUUACGUUUGGGAAUAUGUAAAUAAGCGUCCGUUGUAUUUGUUUUCUUUAGCUUGACGU-- -----------..........((.(((.(((---(.((((((((((..((((((((.(((((((((........))))))))).))).))))).))))))))))))))...)))))..-- ( -27.90, z-score = -3.09, R) >droAna3.scaffold_12916 4711880 110 - 16180835 --------AAACGAAGGACAAAAAUAAAAAGCUCGUGCUGCAAAUAAAAAUGGCGCGUAUUUACGUUUGGGAAUAUGUAAAUAAGCGUCCGUUGUAUUUAUUUUCUUUAGCUUGACGU-- --------..(((.(((...((((((((.(((....))).........((((((((.(((((((((........))))))))).))).)))))...))))))))......)))..)))-- ( -24.60, z-score = -0.98, R) >droWil1.scaffold_180708 2863127 118 - 12563649 GUAUCAAAAUUAAAAAAAGAAGAAAAAAUACACAGUGAAGCAAAUAAAAAUGGUGCGUAUUUACGUUUGGGAAUAUGCAAAUAAGCGUCCGUUGUAUUUAUUUUCUUUAGGUUGAUGU-- .((((((..(.....((((((((..(((((((..(.((.((............((((((((..(....)..)))))))).....)).)))..))))))).)))))))))..)))))).-- ( -21.03, z-score = -1.09, R) >droVir3.scaffold_12963 7172285 88 - 20206255 --------------------------------CAGUCGGGCAAAUAAAAAUGGCGCGUAUUUACGUUUGGGAAUAUGUAAAUAAGAGUCCGUUGUAUUUAUUUCCUUUAGCUUGUGCCGU --------------------------------.....(((.(((((((((((((...(((((((((........)))))))))...).)))))...))))))))))...((....))... ( -18.30, z-score = -0.23, R) >droGri2.scaffold_15252 10319221 88 - 17193109 --------------------------------CAGUCGGGCAAAUAAAAAUGGCGCGUAUUUACGUUUGGGAAUAUGUAAAUAAGAGUCCGUUGUAUUUAUUUCCUUUAGCUUGUGCCAU --------------------------------.....(((.(((((((((((((...(((((((((........)))))))))...).)))))...))))))))))...((....))... ( -18.30, z-score = -0.40, R) >consensus ___________AAAAAAAAAAGUAUAAAAAG___GUGGAGCAAAUAAAAAUGGCGCGUAUUUACGUUUGGGAAUAUGUAAAUAAGCGUCCGUUGUAUUUGUUUUCUUUAGCUUGACGU__ ...................................(((((.(((((((((((((((.(((((((((........))))))))).))).)))))...))))))).)))))........... (-20.11 = -20.31 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:08:03 2011