| Sequence ID | dm3.chr2L |
|---|---|
| Location | 13,443,317 – 13,443,422 |
| Length | 105 |
| Max. P | 0.729345 |

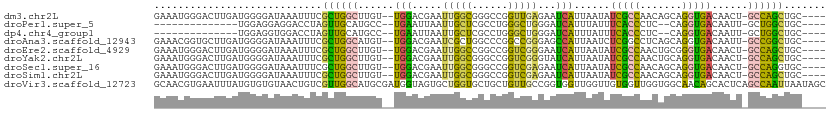
| Location | 13,443,317 – 13,443,422 |
|---|---|
| Length | 105 |
| Sequences | 9 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 67.12 |
| Shannon entropy | 0.65219 |
| G+C content | 0.52379 |
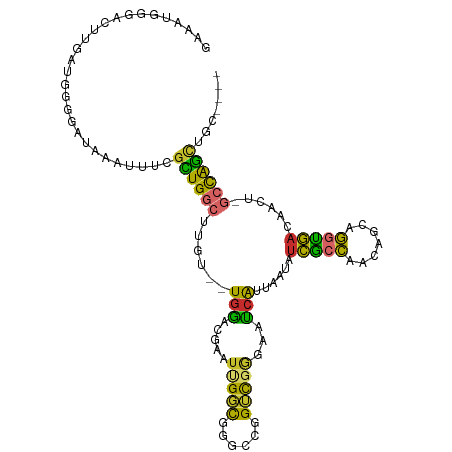
| Mean single sequence MFE | -33.69 |
| Consensus MFE | -14.47 |
| Energy contribution | -13.37 |
| Covariance contribution | -1.10 |
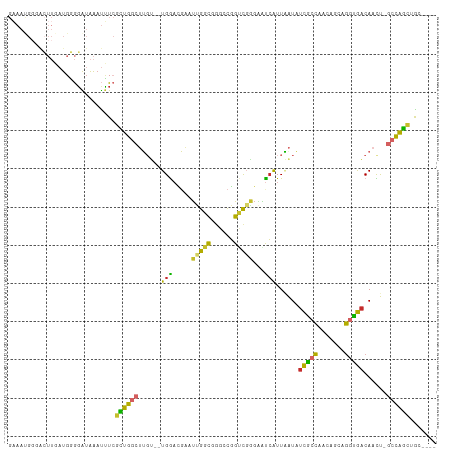
| Combinations/Pair | 1.88 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.729345 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 13443317 105 + 23011544 GAAAUGGGACUUGAUGGGGAUAAAUUUCGCUGGCUUGU--UGGACGAAUUGGCGGGCCGGUUGAGAAUCAUUAAUAUCGCCAACAGCAGGUGACAACU-GCCAGCUGC---- ....(((....(((((..(((.(.((((((((((((((--..(.....)..))))))))).))))).).)))..)))))))).((((.(((.......-))).)))).---- ( -32.50, z-score = -1.49, R) >droPer1.super_5 3120216 89 + 6813705 --------------UGGAGGAGGACCUAGUUGCAUGCC--UGAAUUAAUUGCUCGCCUGGGCUGGGAUCAUUUAUUUCACCCUC--CAGGUGACAAUU-GCUGGCUGC---- --------------...(((....)))....(((.(((--.....((((((.(((((((((..(((.............)))))--))))))))))))-)..))))))---- ( -28.92, z-score = -0.72, R) >dp4.chr4_group1 3173358 89 - 5278887 --------------UGGAGGUGGACCUAGUUGCAUGCC--UGAAUUAAUUGCUCGCCUGGGCUGGGAUCAUUUAUUUCACCCUC--CAGGUGACAAUU-GCUGGCUGC---- --------------...(((....)))....(((.(((--.....((((((.(((((((((..(((.............)))))--))))))))))))-)..))))))---- ( -29.72, z-score = -0.97, R) >droAna3.scaffold_12943 2150080 105 + 5039921 GAAACGGUGCUUGAUGGGGAUAAAUUUCGCUGGCAUGU--UGGACGAAUCGCUGGCCCGGCCGGGAGCCAUUAAUCUCGGCCUCAGCAGGUGACAAUU-GCCGGCUGC---- ((((..((.((.....)).))...))))(((((((.((--((.....((((((((...((((((((........))))))))))))).)))..)))))-))))))...---- ( -39.80, z-score = -1.00, R) >droEre2.scaffold_4929 14642884 105 - 26641161 GAAAUGGGACUUGAUGGGGAUAAAUUUCGCUGGCUUGU--UGGACGAAUUGGCCGGCCGGUCGGGAAUCAUUAAUAUCGCCAACUGCGGGUGACAACU-GCCAGCUGC---- (((((....((.....)).....)))))((((((..((--((...((.((((((....))))))...)).......(((((.......))))))))).-))))))...---- ( -32.10, z-score = -0.69, R) >droYak2.chr2L 9878314 105 + 22324452 GAAAUGGGACUUGAUGGGGAUAAAUUUCGCUGGCUUGU--UGGACGAAUUGGCGGGCCGGUCGGGUAUCAUUAAUAUCGCCAACUGCAGGUGACAACU-GCCAGCUGC---- (((((....((.....)).....)))))((((((..((--((..(((.((((....))))))).............(((((.......))))))))).-))))))...---- ( -31.40, z-score = -0.57, R) >droSec1.super_16 1596726 105 + 1878335 GAAAUGGGACUUGAUGGGGAUAAAUUUCGCUGGCUUGU--UGGACGAAUUGGCGGGCCGGUCGAGAAUCAUUAAUAUCGCCAACAGCAGGUGACAACU-GCCAGGUGC---- ......(.(((((.(((.((((..((((((((((((((--..(.....)..))))))))).)))))........)))).)))...((((.......))-))))))).)---- ( -35.60, z-score = -2.34, R) >droSim1.chr2L 13232966 105 + 22036055 GAAAUGGGACUUGAUGGGGAUAAAUUUCGCUGGCUUGU--UGGACGAAUUGGCGGGCCGGUCGAGAAUCAUUAAUAUCGCCAACAGCAGGUGACAACU-GCCAGCUGC---- ....(((....(((((..(((.(.((((((((((((((--..(.....)..))))))))).))))).).)))..)))))))).((((.(((.......-))).)))).---- ( -34.40, z-score = -1.87, R) >droVir3.scaffold_12723 5269026 112 - 5802038 GCAACGUGAAUUUAUGUGUGUAACUGUCGUUGGCAUGCGAUGGUAGUGCUGGUGCUGCUGUUGCCGGUGGUUGGUUGUGGUUGGUGGCAACAGCACUCAGCCAAUUAAUAGC ((((((((....))))).))).((((((((......)))))))).(.(((((.(.(((((((((((.(..((......))..).)))))))))))))))))).......... ( -38.80, z-score = -0.62, R) >consensus GAAAUGGGACUUGAUGGGGAUAAAUUUCGCUGGCUUGU__UGGACGAAUUGGCGGGCCGGUCGGGAAUCAUUAAUAUCGCCAACAGCAGGUGACAACU_GCCAGCUGC____ ............................((((((..............(((((......)))))............(((((.......)))))......))))))....... (-14.47 = -13.37 + -1.10)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:37:50 2011