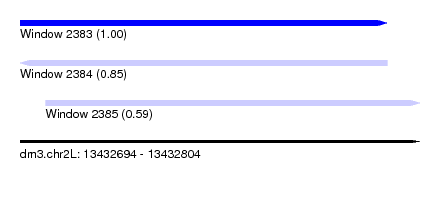
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 13,432,694 – 13,432,804 |
| Length | 110 |
| Max. P | 0.998833 |

| Location | 13,432,694 – 13,432,795 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 79.01 |
| Shannon entropy | 0.38721 |
| G+C content | 0.43048 |
| Mean single sequence MFE | -24.52 |
| Consensus MFE | -15.66 |
| Energy contribution | -17.42 |
| Covariance contribution | 1.76 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.44 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.51 |
| SVM RNA-class probability | 0.998833 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
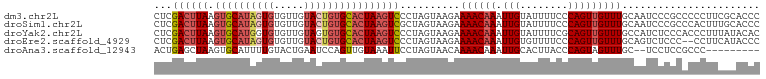
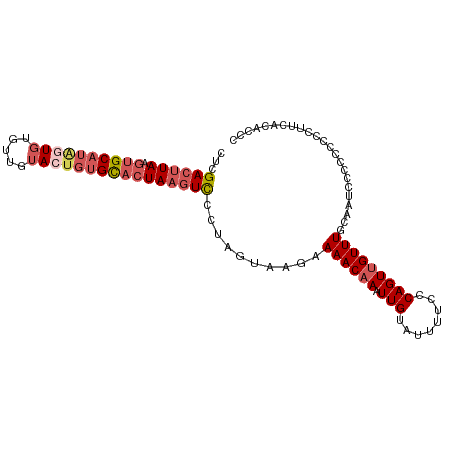
>dm3.chr2L 13432694 101 + 23011544 CUCGACUUAAGUGCAUAGUGUGUUGUACUGUGCACUAAGUCCCUAGUAAGAAAACAAAUUGUAUUUUCCCAGUUGUUUGCAAUCCCGCCCCCUUCGCACCC ...((((((.((((((((((.....))))))))))))))))....(((((.((((((.(((........)))))))))((......))...))).)).... ( -25.50, z-score = -4.37, R) >droSim1.chr2L 13222271 101 + 22036055 CUCGACUUAAGUGCAUAGUGUGUUGUACUGUGCACUAAGUCGCUAGUAAGAAAACAAAUUGUAUUUUCCCAGUUGUUUGCAAUCCCGCCCACUUUGCACCC ..(((((((.((((((((((.....)))))))))))))))))...(((((.((((((.(((........)))))))))((......))....))))).... ( -28.60, z-score = -3.88, R) >droYak2.chr2L 9867765 101 + 22324452 CUCGACUUAAGUGCAUGGUGUGUUGUAGUGUGCACUAAGUCCCUAGUAAGAAAACAAAUUGUAUUUUCGCAGUUGUUUGCCAUCUCCCACCCUUUAUACAC ...((((((.(((((((.((.....)).)))))))))))))....(..(((((((((.((((......))))))))))....)))..)............. ( -21.50, z-score = -1.22, R) >droEre2.scaffold_4929 14632318 99 - 26641161 CUCGACUUAAGUGCAUAGUGUGUUGUACUGUGCACUAAGUCCCUAGUAAGAAAACAAAUUGUGUUUUCCCAGUUGUUUGCAGUCUCCC--CCUUCAUACCC ...((((((.((((((((((.....))))))))))))))))....(((.(((((((.....)))))))..(((((....))).))...--......))).. ( -26.20, z-score = -4.53, R) >droAna3.scaffold_12943 2139161 90 + 5039921 ACUGAGCUAAGUGCAUUUUGUACUGAAUCCAGUUGUAAAUUCCUAGUAACAAAACAAAUUGCACUUACCCAGUAGUUUGC--UCCUCCGCCC--------- ((((.(.((((((((.(((((.(((....)))((((............)))).))))).)))))))).))))).....((--......))..--------- ( -20.80, z-score = -3.19, R) >consensus CUCGACUUAAGUGCAUAGUGUGUUGUACUGUGCACUAAGUCCCUAGUAAGAAAACAAAUUGUAUUUUCCCAGUUGUUUGCAAUCCCCCCCCCUUCACACCC ...((((((.((((((((((.....))))))))))))))))..........((((((.(((........)))))))))....................... (-15.66 = -17.42 + 1.76)
| Location | 13,432,694 – 13,432,795 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 79.01 |
| Shannon entropy | 0.38721 |
| G+C content | 0.43048 |
| Mean single sequence MFE | -26.69 |
| Consensus MFE | -11.84 |
| Energy contribution | -13.64 |
| Covariance contribution | 1.80 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.852597 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 13432694 101 - 23011544 GGGUGCGAAGGGGGCGGGAUUGCAAACAACUGGGAAAAUACAAUUUGUUUUCUUACUAGGGACUUAGUGCACAGUACAACACACUAUGCACUUAAGUCGAG (..(((((...........)))))..)..(((((((((((.....)))))))...)))).(((((((((((.(((.......))).))))).))))))... ( -26.30, z-score = -2.39, R) >droSim1.chr2L 13222271 101 - 22036055 GGGUGCAAAGUGGGCGGGAUUGCAAACAACUGGGAAAAUACAAUUUGUUUUCUUACUAGCGACUUAGUGCACAGUACAACACACUAUGCACUUAAGUCGAG (..(((((...........)))))..)...((((((((((.....))))))))))....((((((((((((.(((.......))).))))).))))))).. ( -28.50, z-score = -2.59, R) >droYak2.chr2L 9867765 101 - 22324452 GUGUAUAAAGGGUGGGAGAUGGCAAACAACUGCGAAAAUACAAUUUGUUUUCUUACUAGGGACUUAGUGCACACUACAACACACCAUGCACUUAAGUCGAG ..........(((((((((..(((......)))......((.....)))))))))))...(((((((((((...............))))).))))))... ( -20.86, z-score = -0.34, R) >droEre2.scaffold_4929 14632318 99 + 26641161 GGGUAUGAAGG--GGGAGACUGCAAACAACUGGGAAAACACAAUUUGUUUUCUUACUAGGGACUUAGUGCACAGUACAACACACUAUGCACUUAAGUCGAG .((((....(.--((....)).).........((((((((.....))))))))))))...(((((((((((.(((.......))).))))).))))))... ( -28.50, z-score = -3.54, R) >droAna3.scaffold_12943 2139161 90 - 5039921 ---------GGGCGGAGGA--GCAAACUACUGGGUAAGUGCAAUUUGUUUUGUUACUAGGAAUUUACAACUGGAUUCAGUACAAAAUGCACUUAGCUCAGU ---------..((......--)).....((((((((((((((.(((((.(((...((((..........))))...))).))))).))))))).))))))) ( -29.30, z-score = -3.89, R) >consensus GGGUACAAAGGGGGGGGGAUUGCAAACAACUGGGAAAAUACAAUUUGUUUUCUUACUAGGGACUUAGUGCACAGUACAACACACUAUGCACUUAAGUCGAG .............(..(((..(((((....((........)).)))))..)))..)....(((((((((((.(((.......))).))))).))))))... (-11.84 = -13.64 + 1.80)


| Location | 13,432,701 – 13,432,804 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 77.18 |
| Shannon entropy | 0.42028 |
| G+C content | 0.41641 |
| Mean single sequence MFE | -20.99 |
| Consensus MFE | -11.54 |
| Energy contribution | -13.06 |
| Covariance contribution | 1.52 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.592799 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 13432701 103 + 23011544 UAAGUGCAUAGUGUGUUGUACUGUGCACUAAGUCCCUAGUAAGAAAACAAAUUGUAUUUUCCCAGUUGUUUGCAAUCCCGCCCCCUUCGCACCCAAAAAUGGC ..(((((((((((.....)))))))))))................................(((.(((..(((...............)))..)))...))). ( -21.56, z-score = -2.09, R) >droSim1.chr2L 13222278 103 + 22036055 UAAGUGCAUAGUGUGUUGUACUGUGCACUAAGUCGCUAGUAAGAAAACAAAUUGUAUUUUCCCAGUUGUUUGCAAUCCCGCCCACUUUGCACCCAAAAAUGGC ..(((((((((((.....))))))))))).....(((((..((((.((.....)).))))..)..(((..(((((...........)))))..)))...)))) ( -24.60, z-score = -1.65, R) >droYak2.chr2L 9867772 103 + 22324452 UAAGUGCAUGGUGUGUUGUAGUGUGCACUAAGUCCCUAGUAAGAAAACAAAUUGUAUUUUCGCAGUUGUUUGCCAUCUCCCACCCUUUAUACACAAAAAUGGC ......(((.((((((...((.(((.((((......))))....((((((.((((......)))))))))).........))).))..))))))....))).. ( -17.90, z-score = 0.64, R) >droEre2.scaffold_4929 14632325 101 - 26641161 UAAGUGCAUAGUGUGUUGUACUGUGCACUAAGUCCCUAGUAAGAAAACAAAUUGUGUUUUCCCAGUUGUUUGCAGUCU--CCCCCUUCAUACCCAAAAAUGGC ..(((((((((((.....))))))))))).............(((((((.....)))))))(((.(((..((.((...--....)).))....)))...))). ( -22.40, z-score = -2.57, R) >droAna3.scaffold_12943 2139168 87 + 5039921 UAAGUGCAUUUUGUACUGAAUCCAGUUGUAAAUUCCUAGUAACAAAACAAAUUGCACUUACCCAGUAGUUUGCUCCUCCGCCCUGGC---------------- ((((((((.(((((.(((....)))((((............)))).))))).)))))))).((((......((......)).)))).---------------- ( -18.50, z-score = -2.14, R) >consensus UAAGUGCAUAGUGUGUUGUACUGUGCACUAAGUCCCUAGUAAGAAAACAAAUUGUAUUUUCCCAGUUGUUUGCAAUCCCGCCCCCUUCACACCCAAAAAUGGC ..(((((((((((.....)))))))))))...............((((((.(((........)))))))))................................ (-11.54 = -13.06 + 1.52)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:37:49 2011