| Sequence ID | dm3.chr2L |
|---|---|
| Location | 13,404,758 – 13,404,865 |
| Length | 107 |
| Max. P | 0.531835 |

| Location | 13,404,758 – 13,404,865 |
|---|---|
| Length | 107 |
| Sequences | 9 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 63.72 |
| Shannon entropy | 0.74096 |
| G+C content | 0.50813 |
| Mean single sequence MFE | -30.57 |
| Consensus MFE | -9.35 |
| Energy contribution | -9.26 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.31 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.531835 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
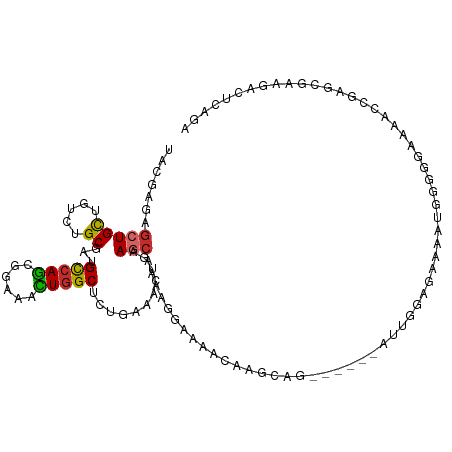
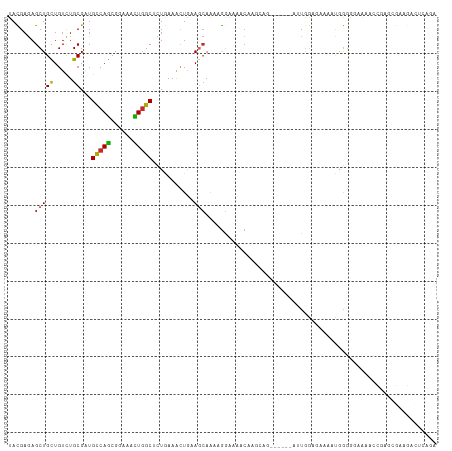
>dm3.chr2L 13404758 107 + 23011544 UACGAGAGCUGCUGUCUGCGAUGCCAGCGGAAACUGGCCCUAAAACUAAAACAAAAGGACAGCAAGCAU------AUUGGAGAGAAUGGGGAAAAACCGAGCGAAGACUCAGA ...(((.(((((((((......(((((......))))).......((........)))))))).)))..------........(..(((.......)))..).....)))... ( -26.60, z-score = -1.32, R) >droSec1.super_16 1563405 107 + 1878335 UACGAGAGCUGCUGUCUGCGAUGCCAGCGGAAACUGGCCCUCAAACUAAAACAAAAGGAUACCAAGCAG------AUUGGAGACAAUGGGGAAAAAUCGAGCGAAGACUCAGA ........(((..((((.(((((((((......)))))((((..............((...))......------((((....))))))))....)))).....)))).))). ( -26.80, z-score = -1.05, R) >droYak2.chr2L 9844198 107 + 22324452 UACGAGAGCUGCUGUCUGCGAUGCCAGCGGAAACUGGCUCUGAAGCUAAAGCAAACGGAAACCAAGCAG------AUUGGAGAAAACGGGGCAAAACCGAGCGAAGACUCAGA ...(((..((.(.((((((...(((((......)))))((((..((....))...))))......))))------)).).))....(((.......)))........)))... ( -30.60, z-score = -0.98, R) >droEre2.scaffold_4929 14608829 107 - 26641161 UACGAGAGCUGCUGUUUGCGAUGUCAGCGGAAACUGGCUCUGAAACUGAAGCAAAAGGAAAGCAAGCAG------GUUGGAGAAAAUGGGACAAAACCGAGCGAAGACUCAGA ...(((.(((.((((((((((.(((((......))))))).....((........))....))))))))------...........(((.......)))))).....)))... ( -25.20, z-score = 0.03, R) >droAna3.scaffold_12943 2109406 107 + 5039921 UAUGAGAGCUGUUGUCUGCGAUGCCAGCGGAAGCUCGCUUUAAAAAUGAAGCAAAAAGAGCCGCGUCGC------GACGGGGAAAAUGGUGGCAAAAAUAGCAAUGAUGCAGA .......(((((((((.(((((((.(((....))).((((((....))))))..........)))))))------)))........((....))..))))))........... ( -31.30, z-score = -0.71, R) >dp4.chr4_group1 3104881 91 - 5278887 UACGAGAGCUGCUGUCUGCGGUGCCAGCGAAAGCUGGCAGUGAAACUGAAGCAGACGGAGAACG--GCAACG---AACAGCAGUCCCAGGAUCAAA----------------- .......((((((((((((...((((((....))))))((.....))...))))))))....((--....))---..)))).(((....)))....----------------- ( -38.90, z-score = -3.98, R) >droPer1.super_5 3076692 91 + 6813705 UACGAGAGCUGCUGUCUGCGGUGCCAGCGAAAGCUGGCUGUGAAACUGAAGCAGACGGAGAACG--GCAACG---AACAGCAGUCCCAGGAUCAAA----------------- .......((((((((((((...((((((....)))))).(.....)....))))))))....((--....))---..)))).(((....)))....----------------- ( -37.00, z-score = -3.18, R) >droWil1.scaffold_180708 7208109 110 - 12563649 UAUGAAAGCUGUUGUCUGCGAUGCCAACGUAAAUUGGCCUUGACACUGAAGAAAAAGCAAGACAAUCAGUCCAAUGUUGAAUCCAAGGAGAGAAAACCGACAGAUGACGA--- .........(((..((((((..(((((......)))))(((....((...((...((((.(((.....)))...))))...))..))..))).....)).))))..))).--- ( -23.00, z-score = -1.00, R) >droGri2.scaffold_15252 13037017 104 - 17193109 UACGAGAGCUGCUGUCUGCGCUGCCAGCGCAAAUUGGCACUGAAAAUGAAGCAGCAGAGCAGCGAAGAAACG---GCUGACUCUACUAGUGGUGGUGCUGCUGCUGC------ ......((.(((((..(((((.....)))))...))))))).........((((((..((((((.....((.---((((.......)))).))..))))))))))))------ ( -35.70, z-score = 0.49, R) >consensus UACGAGAGCUGCUGUCUGCGAUGCCAGCGGAAACUGGCUCUGAAACUGAAGCAAAAGGAAAACAAGCAG______AUUGGAGAAAAUGGGGGAAAACCGAGCGAAGACUCAGA .......(((((.....))...(((((......)))))...........)))............................................................. ( -9.35 = -9.26 + -0.10)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:37:43 2011