
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 13,402,293 – 13,402,401 |
| Length | 108 |
| Max. P | 0.843280 |

| Location | 13,402,293 – 13,402,401 |
|---|---|
| Length | 108 |
| Sequences | 9 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 72.27 |
| Shannon entropy | 0.53753 |
| G+C content | 0.38016 |
| Mean single sequence MFE | -23.28 |
| Consensus MFE | -8.27 |
| Energy contribution | -7.44 |
| Covariance contribution | -0.82 |
| Combinations/Pair | 1.41 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.843280 |
| Prediction | RNA |
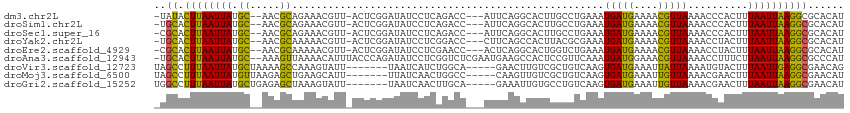
Download alignment: ClustalW | MAF
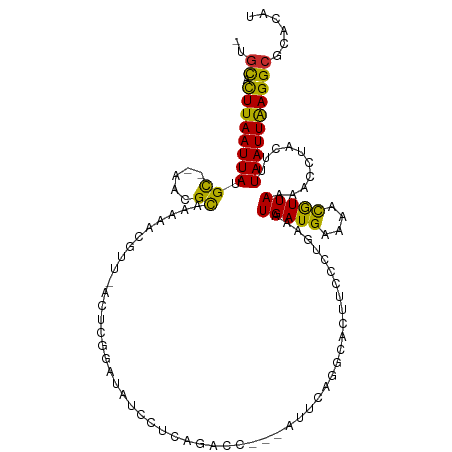
>dm3.chr2L 13402293 108 + 23011544 -UAUACUUAAUUAUGC--AACGCAGAAACGUU-ACUCGGAUAUCCUCAGACC---AUUCAGGCACUUGCCUGAAAUGAUGAAAACGUUAAAACCCACUUUAAUUAAGGCGCACAU -....(((((((((((--...)))..((((((-....((....))(((...(---((((((((....))))).)))).))).))))))...........))))))))........ ( -25.10, z-score = -3.36, R) >droSim1.chr2L 13196506 108 + 22036055 -UGCACUUAAUUAUGC--AACGCAGAAACGUU-ACUCGGAUAUCCUCAGACC---AUUCAGGCACUUGCCUGAAAUGAUGAAAACGUUAAAACCCACUUUAAUUAAGGCGCACAU -(((.(((((((((((--...)))..((((((-....((....))(((...(---((((((((....))))).)))).))).))))))...........))))))))..)))... ( -28.20, z-score = -3.85, R) >droSec1.super_16 1560892 108 + 1878335 -CGCACUUAAUUAUGC--AACGCAGAAACGUU-ACUCGGAUAUCCUCAGACC---AUUCAGGCACUUGCCUGAAAUGAUGAAAACGUUAAAACCCACUUUAAUUAAGGCGCACAU -(((.(((((((((((--...)))..((((((-....((....))(((...(---((((((((....))))).)))).))).))))))...........)))))))))))..... ( -29.00, z-score = -4.18, R) >droYak2.chr2L 9841712 108 + 22324452 -UGCACUUAAUUAUGC--AACGCAAAAACGUU-ACUCGGAUAUCCUCGGACC---CUUCAGCCACUUACGCGAAAUGAUGAAAACGUUAAAACCUACUUUAAUUAAGGCGCACAU -(((.(((((((((((--...)))..((((((-....((....))(((....---.(((.((.......)))))....))).))))))...........))))))))..)))... ( -17.10, z-score = -0.41, R) >droEre2.scaffold_4929 14606341 108 - 26641161 -CGCACUUAAUUAUGC--AACGCAAAAACGUU-ACUCGGAUAUCCUCGAACC---ACUCAGGCACUGGUCUGAAAUGAUGAAAACGUUAAAACCUACUUUAAUUAAGGCGCACAU -(((.(((((((((((--...)))..((((((-..((((......))))...---..((((((....)))))).........))))))...........)))))))))))..... ( -23.90, z-score = -2.65, R) >droAna3.scaffold_12943 2107012 112 + 5039921 -UGCACUUAAUUAUGC--AAAGUUAAAACAUUUACCCAGAUAUCCUCGGUCUCGAAUGAAGCCACUCCGUUCAAAUGAUGGAAACGUUAAAACCUUUCUUAAUUAAGGCGCCCAU -.((.((((((((.(.--((((...........(((.((.....)).)))...(((((.((...)).)))))...(((((....)))))....))))).))))))))))...... ( -21.50, z-score = -1.63, R) >droVir3.scaffold_12723 5195206 103 - 5802038 UAGCCUUUAAUUAUGCUAAAAGCCAAAGUAUU-------UAAUCAUCUGGCA-----GAACUUGUCGCUGUCAAGUGAUGAAAUUAUUAAAAUGUACUUUAAUUGAGGCGAACAG ..(((((.((((..((.....)).(((((((.-------..((((..(((((-----(.........))))))..))))..............))))))))))))))))...... ( -22.43, z-score = -1.67, R) >droMoj3.scaffold_6500 3232404 103 - 32352404 UAGCCUUUAAUUAUGUUAAGAGCUGAAGCAUU-------UUAUCAACUGGCC-----CAAGUUGUCGCUGUCAAGUGAUGAAAUUGUUAAAACGAACUUUAAUUAAGGCGAACAU ..(((((.(((((.(((....((....))(((-------((((((..((((.-----..(((....)))))))..)))))))))..........)))..))))))))))...... ( -22.60, z-score = -1.35, R) >droGri2.scaffold_15252 13034761 103 - 17193109 UGGCCUUUAAUUAUGCUGAGAGCUAAAGUAUU-------UAAUCAACUUGCA-----GAAAUUGUGCCUGUCAAGUGAUGAAAUUGUUAAAACGAACUUUAAUUAAGGCGAACAU ..(((((.......((.....))((((((...-------..((((.((((((-----(.........))).))))))))....((((....))))))))))...)))))...... ( -19.70, z-score = -0.74, R) >consensus _UGCACUUAAUUAUGC__AACGCAAAAACGUU_ACUCGGAUAUCCUCAGACC___AUUCAGGCACUUCCCUGAAAUGAUGAAAACGUUAAAACCUACUUUAAUUAAGGCGCACAU ..((.((((((((.((.....))....................................................(((((....)))))..........))))))))))...... ( -8.27 = -7.44 + -0.82)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:37:42 2011