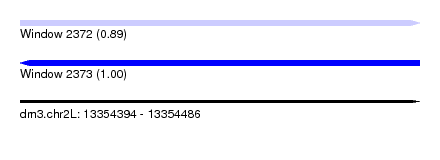
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 13,354,394 – 13,354,486 |
| Length | 92 |
| Max. P | 0.996867 |

| Location | 13,354,394 – 13,354,486 |
|---|---|
| Length | 92 |
| Sequences | 12 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 70.10 |
| Shannon entropy | 0.59375 |
| G+C content | 0.62050 |
| Mean single sequence MFE | -39.45 |
| Consensus MFE | -23.35 |
| Energy contribution | -23.35 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.97 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.885295 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
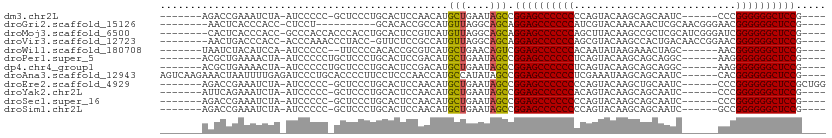
>dm3.chr2L 13354394 92 + 23011544 ----CGGAGCCCCCCGGG------GAUUGCUGCUUGUACUGGGGGGGGCUCCGGCUAUUCAGCAUGUUGGAGUGCAGGGAGC-GGGGGAU-UAGAUUUCGGUCU------- ----(((((((((((..(------(..(((.....)))))..)))))))))))((..(((((....)))))..))..(((.(-(..(...-....)..)).)))------- ( -40.00, z-score = -1.24, R) >droGri2.scaffold_15126 56577 88 + 8399593 ----CGGAGCCCCCCGUUCCCGUUGCGAGUUGUUUGUACGAUGGGGGGCUCCUGCUGCCUAACAUGGCGGUGUGC----------AGGAG-GGUGGGUGAGUU-------- ----.((((((((((((...(((.(((((...)))))))))))))))))))).((((((......))))))....----------.....-............-------- ( -39.40, z-score = -1.70, R) >droMoj3.scaffold_6500 6372101 98 + 32352404 ----CGGAGCCCCCCGAUCCCGAUGCGAGCGGCUUGUAAGCUGGGGGGCUCCUGCUGCCUAACAUGACGGAGUGCAGGUGGGUGGUGGGC-GGUGGGUGAGUG-------- ----.(((((((((((.(..(((.((.....)))))..)..))))))))))).((((((((.(((..(........)....))).)))))-))).........-------- ( -41.10, z-score = 0.06, R) >droVir3.scaffold_12723 2393604 97 - 5802038 ----CGGAGCCCCCCGUUCCGGUUGUCAGUGGCUUGUACGCUGGGGGGCUCCUGCUGCCUAACAUGGCGGAGAAC-GGUAGGGUUUGGGU-GGUGGGUCAGUU-------- ----.((((((((((((.(.((((......)))).)...)).)))))))))).((..((((((.((.((.....)-).))..).))))).-.)).........-------- ( -39.90, z-score = -0.12, R) >droWil1.scaffold_180708 7131544 91 - 12563649 ----CGGAGCCCCCCGUU------GCUAGUUUCUUAUAUUGUGGGGGGCUCCGACUGUUCAGCAUGACGCGGUGUGGGGAA--GGGGGAU-UGGAUGUAGAUUA------- ----((((((((((((..------..((((.......)))))))))))))))).((.(((.((((......))))...)))--.))....-.............------- ( -33.20, z-score = -1.15, R) >droPer1.super_5 3013992 93 + 6813705 ----CGGAGCCCCCCCUU------GCCUGCUGCUUGUACUGAGGGGGGCUCCGGCUAUUCAGCAUGUCGGAGUGCAGGGAGCAGGGGGAU-UAGUUUUCAGCGU------- ----(((((((((((.((------...(((.....)))..)))))))))))))(((.....((((......)))).(..(((..(....)-..)))..))))..------- ( -38.10, z-score = 0.00, R) >dp4.chr4_group1 3043228 93 - 5278887 ----CGGAGCCCCCCCUU------GCCUGCUGCUUGUACUGAGGGGGGCUCCGGCUAUUCAGCAUGUCGGAGUGCAGGGAGCAGGGGGAU-UAGUUUUCAGCGU------- ----(((((((((((.((------...(((.....)))..)))))))))))))(((.....((((......)))).(..(((..(....)-..)))..))))..------- ( -38.10, z-score = 0.00, R) >droAna3.scaffold_12943 2005140 101 + 5039921 ----CGGAGCCCCCCGUG------GAUUGCUGCUUAUUUCGAGGGGGGCUCCGGCUAUAUGGCAUGGUUGGGAGGAAGGGGUGCAGGGAUCUCAAAAUUAGUUUCUUGACU ----(((((((((((.((------((...........)))).)))))))))))(((....)))..(((..(((((..(((((......)))))........)))))..))) ( -40.30, z-score = -1.59, R) >droEre2.scaffold_4929 14554403 96 - 26641161 CCAGCGGAGCCCCCCGGG------GAUUGCAGCUUGUACUGGGGGGGGCUCCGGCUAUUCAGCAUGUUGGAGUGCAGGGAGC-GGGGGAU-UAGAUUUCGGUCU------- (((((((((((((((..(------(..(((.....)))))..)))))))))).(((....)))..))))).......(((.(-(..(...-....)..)).)))------- ( -42.00, z-score = -1.16, R) >droYak2.chr2L 9792868 92 + 22324452 ----CGGAGCCCCCCGGG------GAUUGCUGCUUGUACUGUGGGGGGCUCCGGCUAUUCAGCAUGUUGGAGUGCAGGGAGC-GGGGGAU-UAGAUUUCUGAAU------- ----((((((((((((.(------(..(((.....))))).))))))))))))(((.(((.((((......)))).))))))-......(-(((....))))..------- ( -41.30, z-score = -2.36, R) >droSec1.super_16 1512834 92 + 1878335 ----CGGAGCCCCCCGGG------GAUUGCUGCUUGUACUGGGGGGGGCUCCGGCUAUUCAGCAUGUUGGAGUGCAGGGAGC-GGGGGAU-UAGAUUUCGGUCU------- ----(((((((((((..(------(..(((.....)))))..)))))))))))((..(((((....)))))..))..(((.(-(..(...-....)..)).)))------- ( -40.00, z-score = -1.24, R) >droSim1.chr2L 13142595 92 + 22036055 ----CGGAGCCCCCCGGC------GAUUGCUGCUUGUACUGGGGGGGGCUCCGGCUAUUCAGCAUGUUGGAGUGCAGGGAGC-GGGGGAU-UAGAUUUCGGUCU------- ----((((((((((((((------....))).((......)))))))))))))((..(((((....)))))..))..(((.(-(..(...-....)..)).)))------- ( -40.00, z-score = -1.16, R) >consensus ____CGGAGCCCCCCGUU______GAUUGCUGCUUGUACUGUGGGGGGCUCCGGCUAUUCAGCAUGUCGGAGUGCAGGGAGC_GGGGGAU_UAGAUUUCAGUCU_______ .....((((((((((...........................))))))))))........................................................... (-23.35 = -23.35 + 0.00)

| Location | 13,354,394 – 13,354,486 |
|---|---|
| Length | 92 |
| Sequences | 12 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 70.10 |
| Shannon entropy | 0.59375 |
| G+C content | 0.62050 |
| Mean single sequence MFE | -30.11 |
| Consensus MFE | -23.91 |
| Energy contribution | -23.73 |
| Covariance contribution | -0.18 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.00 |
| SVM RNA-class probability | 0.996867 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 13354394 92 - 23011544 -------AGACCGAAAUCUA-AUCCCCC-GCUCCCUGCACUCCAACAUGCUGAAUAGCCGGAGCCCCCCCCAGUACAAGCAGCAAUC------CCCGGGGGGCUCCG---- -------.............-.......-(((....(((........))).....)))(((((((((((..................------...)))))))))))---- ( -27.70, z-score = -1.82, R) >droGri2.scaffold_15126 56577 88 - 8399593 --------AACUCACCCACC-CUCCU----------GCACACCGCCAUGUUAGGCAGCAGGAGCCCCCCAUCGUACAAACAACUCGCAACGGGAACGGGGGGCUCCG---- --------............-.....----------.......(((......)))....((((((((((...((....))..((((...))))...)))))))))).---- ( -32.80, z-score = -2.64, R) >droMoj3.scaffold_6500 6372101 98 - 32352404 --------CACUCACCCACC-GCCCACCACCCACCUGCACUCCGUCAUGUUAGGCAGCAGGAGCCCCCCAGCUUACAAGCCGCUCGCAUCGGGAUCGGGGGGCUCCG---- --------............-............((((.((........)))))).....((((((((((.(((....)))..((((...))))...)))))))))).---- ( -33.20, z-score = -1.02, R) >droVir3.scaffold_12723 2393604 97 + 5802038 --------AACUGACCCACC-ACCCAAACCCUACC-GUUCUCCGCCAUGUUAGGCAGCAGGAGCCCCCCAGCGUACAAGCCACUGACAACCGGAACGGGGGGCUCCG---- --------............-..............-.......(((......)))....((((((((((.((......))..(((.....)))...)))))))))).---- ( -34.00, z-score = -2.15, R) >droWil1.scaffold_180708 7131544 91 + 12563649 -------UAAUCUACAUCCA-AUCCCCC--UUCCCCACACCGCGUCAUGCUGAACAGUCGGAGCCCCCCACAAUAUAAGAAACUAGC------AACGGGGGGCUCCG---- -------.............-.......--...........((.....))........(((((((((((..................------...)))))))))))---- ( -24.80, z-score = -2.46, R) >droPer1.super_5 3013992 93 - 6813705 -------ACGCUGAAAACUA-AUCCCCCUGCUCCCUGCACUCCGACAUGCUGAAUAGCCGGAGCCCCCCUCAGUACAAGCAGCAGGC------AAGGGGGGGCUCCG---- -------..((((.......-.......(((.....)))...............))))((((((((((((..((...........))------..))))))))))))---- ( -31.35, z-score = -0.52, R) >dp4.chr4_group1 3043228 93 + 5278887 -------ACGCUGAAAACUA-AUCCCCCUGCUCCCUGCACUCCGACAUGCUGAAUAGCCGGAGCCCCCCUCAGUACAAGCAGCAGGC------AAGGGGGGGCUCCG---- -------..((((.......-.......(((.....)))...............))))((((((((((((..((...........))------..))))))))))))---- ( -31.35, z-score = -0.52, R) >droAna3.scaffold_12943 2005140 101 - 5039921 AGUCAAGAAACUAAUUUUGAGAUCCCUGCACCCCUUCCUCCCAACCAUGCCAUAUAGCCGGAGCCCCCCUCGAAAUAAGCAGCAAUC------CACGGGGGGCUCCG---- ..((((((......))))))......................................(((((((((((..................------...)))))))))))---- ( -27.20, z-score = -1.70, R) >droEre2.scaffold_4929 14554403 96 + 26641161 -------AGACCGAAAUCUA-AUCCCCC-GCUCCCUGCACUCCAACAUGCUGAAUAGCCGGAGCCCCCCCCAGUACAAGCUGCAAUC------CCCGGGGGGCUCCGCUGG -------...(((.......-.......-(((....(((........))).....)))(((((((((((.((((....)))).....------...))))))))))).))) ( -31.70, z-score = -1.82, R) >droYak2.chr2L 9792868 92 - 22324452 -------AUUCAGAAAUCUA-AUCCCCC-GCUCCCUGCACUCCAACAUGCUGAAUAGCCGGAGCCCCCCACAGUACAAGCAGCAAUC------CCCGGGGGGCUCCG---- -------((((((.......-.......-((.....))...........))))))...(((((((((((..................------...)))))))))))---- ( -29.31, z-score = -2.68, R) >droSec1.super_16 1512834 92 - 1878335 -------AGACCGAAAUCUA-AUCCCCC-GCUCCCUGCACUCCAACAUGCUGAAUAGCCGGAGCCCCCCCCAGUACAAGCAGCAAUC------CCCGGGGGGCUCCG---- -------.............-.......-(((....(((........))).....)))(((((((((((..................------...)))))))))))---- ( -27.70, z-score = -1.82, R) >droSim1.chr2L 13142595 92 - 22036055 -------AGACCGAAAUCUA-AUCCCCC-GCUCCCUGCACUCCAACAUGCUGAAUAGCCGGAGCCCCCCCCAGUACAAGCAGCAAUC------GCCGGGGGGCUCCG---- -------.............-.......-(((....(((........))).....)))(((((((((((...((....)).((....------)).)))))))))))---- ( -30.20, z-score = -2.22, R) >consensus _______AAACUGAAAUCUA_AUCCCCC_GCUCCCUGCACUCCAACAUGCUGAAUAGCCGGAGCCCCCCACAGUACAAGCAGCAAUC______AACGGGGGGCUCCG____ ................................................(((....))).((((((((((...........................))))))))))..... (-23.91 = -23.73 + -0.18)
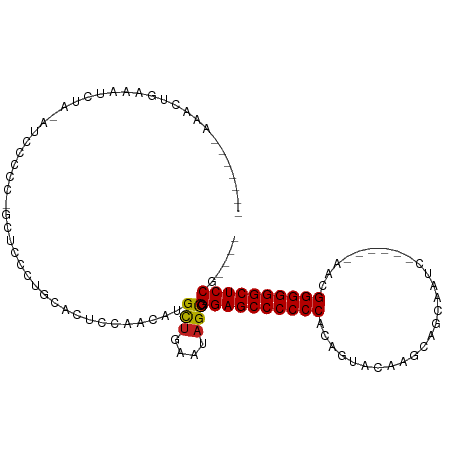
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:37:39 2011