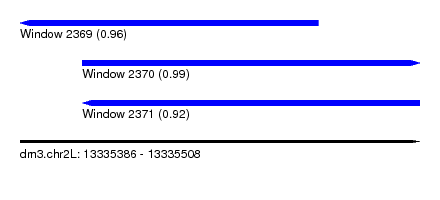
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 13,335,386 – 13,335,508 |
| Length | 122 |
| Max. P | 0.986044 |

| Location | 13,335,386 – 13,335,477 |
|---|---|
| Length | 91 |
| Sequences | 10 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 78.98 |
| Shannon entropy | 0.40297 |
| G+C content | 0.35601 |
| Mean single sequence MFE | -23.51 |
| Consensus MFE | -16.84 |
| Energy contribution | -16.20 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.46 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.964593 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 13335386 91 - 23011544 CUGUGUGUCUAACGUUUUGUUGAUUGAUGGUUAACCAGAAAAUCAAUAUCACUAUGCUAAUGAGACU-GGCAUAAAUGCACAAACAUAUGUA--------------------- .(((((((.....((..((((((((..(((....)))...))))))))..))(((((((.......)-)))))).)))))))..........--------------------- ( -23.70, z-score = -2.51, R) >droAna3.scaffold_12943 1979566 112 - 5039921 UUGUGUGUCCAACGUUUUGUUGAUUGACGGUUAGCCGGAAAAUCAAUAUCACUAUGCUAAUGAGACU-CACAUAAGUGCACAAACAUAUGUACACAGAUAUGUAAGUUUGGGU .......(((...((..((((((((..(((....)))...))))))))..))..........(((((-.(((((.(((.(((......))).)))...))))).)))))))). ( -26.30, z-score = -0.41, R) >droEre2.scaffold_4929 14535376 91 + 26641161 CUGUGUGUCUAACGUUUUGUUGAUUGAUGGUUAACCGGAAAAUCAAUAUCACUAUGCUAAUGAGACU-GGCAUAAAUGCACAAACAUAUGUA--------------------- .(((((((.....((..((((((((..(((....)))...))))))))..))(((((((.......)-)))))).)))))))..........--------------------- ( -23.00, z-score = -2.31, R) >droYak2.chr2L 9772576 91 - 22324452 CUGUGUGUCUAACGUUUUGUUGAUUGAUGGUUAACCAGAAAAUCAAUAUCACUAUGCUAAUGAGACU-GGCAUAGAUGCACAAACAUAUGUG--------------------- .((((((((((..((..((((((((..(((....)))...))))))))..))..(((((.......)-))))))))))))))..........--------------------- ( -27.80, z-score = -3.57, R) >droSec1.super_16 1493956 91 - 1878335 CUGUGUGUCUAACGUUUUGUUGAUUGAUGGUUAACCAGAAAAUCAAUAUCACUAUGCUAAUGAGACU-GGCAUAGAUGUACAAACAUAUGUA--------------------- .((((..((((..((..((((((((..(((....)))...))))))))..))..(((((.......)-))))))))..))))..........--------------------- ( -25.20, z-score = -2.82, R) >droSim1.chr2L 13120341 91 - 22036055 CUGUGUGUCUAACGUUUUGUUGAUUGAUGGUUAACCAGAAAAUCAAUAUCACUAUGCUAAUGAGACU-GGCAUAAAUGUACAAACAUAUGUA--------------------- ..((((((...(((((.((((((((..(((....)))...))))))))....(((((((.......)-)))))))))))....))))))...--------------------- ( -21.90, z-score = -2.02, R) >droPer1.super_5 4699452 91 - 6813705 AUGUGUGAGUGGCGUUUUGUUGAUUGAUGGUUAACCGGAAAAUCAAUAUCACUAUGCUAAUGAGACU-AGCACACUCGUCUAUGUAUGCGUA--------------------- (((..((((((..((..((((((((..(((....)))...))))))))..))..(((((.......)-))))))))))..))).........--------------------- ( -23.80, z-score = -1.25, R) >dp4.chr4_group5 198986 91 - 2436548 AUGUGUGAGUGGCGUUUUGUUGAUUGAUGGUUAACCGGAAAAUCAAUAUCACUAUGCUAAUGAGACU-AGCAUACUCGUCUAUGUAUGCGUA--------------------- ..((.(.(.((((((..((((((((..(((....)))...)))))))).....)))))).).).)).-.((((((........))))))...--------------------- ( -25.80, z-score = -2.22, R) >droWil1.scaffold_180708 7076851 92 + 12563649 AUAUAUGUAUACCGUUUUGUUGAUUGAUGGUUAACCAGAAAAUCAAUAUCACUAUGCUAAUGAGACUAGGCAUAAAUGCACAUACACAGACA--------------------- .(((.(((((...((..((((((((..(((....)))...))))))))..))((((((..........)))))).))))).)))........--------------------- ( -20.00, z-score = -1.83, R) >droGri2.scaffold_15126 32441 99 - 8399593 UUUUGUGUCUGUCGUUUUGUUGAUUGAUGGUUAACCAGGAAAUCAAUAUUGCAAUGCUAAUGAGAAUGAAAACGAGAACAAGAAGAAGAACUGCAACGA-------------- ((((((.((..((((((((((((((..(((....)))...)))))))...((...))...........)))))))))))))))................-------------- ( -17.60, z-score = 0.00, R) >consensus CUGUGUGUCUAACGUUUUGUUGAUUGAUGGUUAACCAGAAAAUCAAUAUCACUAUGCUAAUGAGACU_GGCAUAAAUGCACAAACAUAUGUA_____________________ .(((((((.....((..((((((((..(((....)))...))))))))..))((((((..........)))))).)))))))............................... (-16.84 = -16.20 + -0.64)

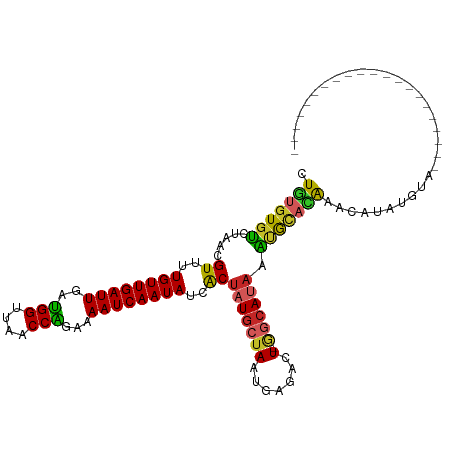
| Location | 13,335,405 – 13,335,508 |
|---|---|
| Length | 103 |
| Sequences | 11 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 84.53 |
| Shannon entropy | 0.31091 |
| G+C content | 0.38429 |
| Mean single sequence MFE | -27.38 |
| Consensus MFE | -20.21 |
| Energy contribution | -20.50 |
| Covariance contribution | 0.29 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.22 |
| SVM RNA-class probability | 0.986044 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 13335405 103 + 23011544 ------AUGCC-AGUCUCAUUAGCAUAGUGAUAUUGAUUUUCUGGUUAACCAUCAAUCAACAAAACGUUAGACACACAGGCCCCAGAU---------UGCUAUGCAAUUGAGGCUCCAA ------.....-(((((((...((((((..((.((((((...(((....)))..))))))..................(....)..))---------..))))))...))))))).... ( -26.50, z-score = -2.33, R) >droAna3.scaffold_12943 1979606 105 + 5039921 ------AUGUG-AGUCUCAUUAGCAUAGUGAUAUUGAUUUUCCGGCUAACCGUCAAUCAACAAAACGUUGGACACACAAACCAAAGAUA-------UUGCUAUGCAAUUGAGGCUACAA ------.(((.-(((((((...((((((..(((((((((...(((....)))..)))).........((((.........)))).))))-------)..))))))...)))))))))). ( -33.50, z-score = -4.70, R) >droEre2.scaffold_4929 14535395 103 - 26641161 ------AUGCC-AGUCUCAUUAGCAUAGUGAUAUUGAUUUUCCGGUUAACCAUCAAUCAACAAAACGUUAGACACACAGGCCCCAGAU---------UGCUAUGCAAUUGAGGCUCCAA ------.....-(((((((...((((((..((.(((.....((((....)).....(((((.....))).))......))...)))))---------..))))))...))))))).... ( -24.70, z-score = -2.09, R) >droYak2.chr2L 9772595 103 + 22324452 ------AUGCC-AGUCUCAUUAGCAUAGUGAUAUUGAUUUUCUGGUUAACCAUCAAUCAACAAAACGUUAGACACACAGGCCCCAGAU---------UGCUAUGCAAUUGAGGCUCCAA ------.....-(((((((...((((((..((.((((((...(((....)))..))))))..................(....)..))---------..))))))...))))))).... ( -26.50, z-score = -2.33, R) >droSec1.super_16 1493975 103 + 1878335 ------AUGCC-AGUCUCAUUAGCAUAGUGAUAUUGAUUUUCUGGUUAACCAUCAAUCAACAAAACGUUAGACACACAGGCCCCAGAU---------UGCUAUGCAAUUGAGGCUCCAA ------.....-(((((((...((((((..((.((((((...(((....)))..))))))..................(....)..))---------..))))))...))))))).... ( -26.50, z-score = -2.33, R) >droSim1.chr2L 13120360 103 + 22036055 ------AUGCC-AGUCUCAUUAGCAUAGUGAUAUUGAUUUUCUGGUUAACCAUCAAUCAACAAAACGUUAGACACACAGGCCCCAGAU---------UGCUAUGCAAUUGAGGCUCCAA ------.....-(((((((...((((((..((.((((((...(((....)))..))))))..................(....)..))---------..))))))...))))))).... ( -26.50, z-score = -2.33, R) >droPer1.super_5 4699471 112 + 6813705 ------GUGCU-AGUCUCAUUAGCAUAGUGAUAUUGAUUUUCCGGUUAACCAUCAAUCAACAAAACGCCACUCACACAUGUAUGAGAUGUAUGAGAUUGCUAUGCAAUUGAGGCUUCAA ------.((..-(((((((...((((((..((.((((((....((....))...))))))..........((((.((((.......)))).))))))..))))))...))))))).)). ( -32.80, z-score = -3.04, R) >dp4.chr4_group5 199005 112 + 2436548 ------AUGCU-AGUCUCAUUAGCAUAGUGAUAUUGAUUUUCCGGUUAACCAUCAAUCAACAAAACGCCACUCACACAUGUAUGAGAUGUAUGAGAUUGCUAUGCAAUUGAGGCUUCAA ------.((..-(((((((...((((((..((.((((((....((....))...))))))..........((((.((((.......)))).))))))..))))))...))))))).)). ( -32.90, z-score = -3.26, R) >droWil1.scaffold_180708 7076870 109 - 12563649 ------AUGCCUAGUCUCAUUAGCAUAGUGAUAUUGAUUUUCUGGUUAACCAUCAAUCAACAAAACGGUAUACAUAUAUGUACAAAAUGA----GAUUGCUAUGCAAUUGAGGCUCCAA ------......(((((((...((((((..((.((((((...(((....)))..)))))).......(((((......))))).......----.))..))))))...))))))).... ( -29.50, z-score = -3.09, R) >droVir3.scaffold_12723 2362495 99 - 5802038 ------GUGCU-UGUCUCAUUAGCAUAGUGAUAUUGAUUUUCUG-UUAACCAUCAAUCAACAAAACGCCAUACACAAAA---UGAGAU---------UGCAAUGCAAUUGAGGCUUCAA ------.....-.((((((...((((.(..((.((.(((((.((-(.........................))).))))---).))))---------..).))))...))))))..... ( -18.51, z-score = -0.14, R) >droGri2.scaffold_15126 32461 107 + 8399593 GUUCUCGUUUUCAUUCUCAUUAGCAUUGCAAUAUUGAUUUCCUGGUUAACCAUCAAUCAACAAAACGACAGACACAAAA---UUAGAU---------UGCAAUGCAAUUGAGGCUUCAA ..............(((((...((((((((((.((((((...(((....)))..)))))).....(....)........---....))---------))))))))...)))))...... ( -23.30, z-score = -2.62, R) >consensus ______AUGCC_AGUCUCAUUAGCAUAGUGAUAUUGAUUUUCUGGUUAACCAUCAAUCAACAAAACGUUAGACACACAGGCCCCAGAU_________UGCUAUGCAAUUGAGGCUCCAA ............(((((((...(((((((....((((((...(((....)))..))))))......(.....).........................)))))))...))))))).... (-20.21 = -20.50 + 0.29)

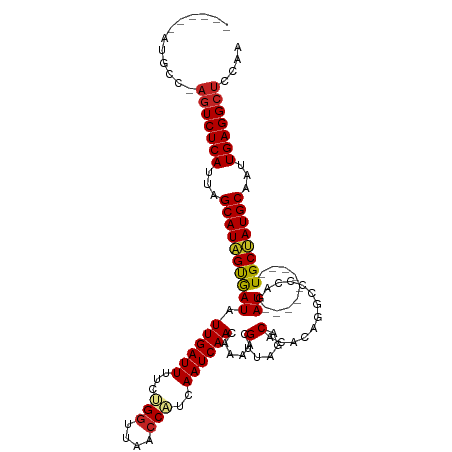
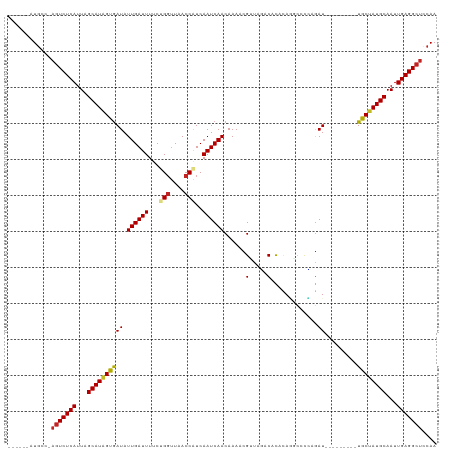
| Location | 13,335,405 – 13,335,508 |
|---|---|
| Length | 103 |
| Sequences | 11 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 84.53 |
| Shannon entropy | 0.31091 |
| G+C content | 0.38429 |
| Mean single sequence MFE | -29.42 |
| Consensus MFE | -20.55 |
| Energy contribution | -20.39 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.918400 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 13335405 103 - 23011544 UUGGAGCCUCAAUUGCAUAGCA---------AUCUGGGGCCUGUGUGUCUAACGUUUUGUUGAUUGAUGGUUAACCAGAAAAUCAAUAUCACUAUGCUAAUGAGACU-GGCAU------ ..((.(((((((((((...)))---------)).))))))))...........((..((((((((..(((....)))...))))))))..)).((((((.......)-)))))------ ( -29.30, z-score = -1.69, R) >droAna3.scaffold_12943 1979606 105 - 5039921 UUGUAGCCUCAAUUGCAUAGCAA-------UAUCUUUGGUUUGUGUGUCCAACGUUUUGUUGAUUGACGGUUAGCCGGAAAAUCAAUAUCACUAUGCUAAUGAGACU-CACAU------ .(((((.((((...((((((...-------.....((((.........)))).....((((((((..(((....)))...))))))))...))))))...)))).))-.))).------ ( -27.90, z-score = -2.01, R) >droEre2.scaffold_4929 14535395 103 + 26641161 UUGGAGCCUCAAUUGCAUAGCA---------AUCUGGGGCCUGUGUGUCUAACGUUUUGUUGAUUGAUGGUUAACCGGAAAAUCAAUAUCACUAUGCUAAUGAGACU-GGCAU------ ..((.(((((((((((...)))---------)).))))))))...........((..((((((((..(((....)))...))))))))..)).((((((.......)-)))))------ ( -28.60, z-score = -1.45, R) >droYak2.chr2L 9772595 103 - 22324452 UUGGAGCCUCAAUUGCAUAGCA---------AUCUGGGGCCUGUGUGUCUAACGUUUUGUUGAUUGAUGGUUAACCAGAAAAUCAAUAUCACUAUGCUAAUGAGACU-GGCAU------ ..((.(((((((((((...)))---------)).))))))))...........((..((((((((..(((....)))...))))))))..)).((((((.......)-)))))------ ( -29.30, z-score = -1.69, R) >droSec1.super_16 1493975 103 - 1878335 UUGGAGCCUCAAUUGCAUAGCA---------AUCUGGGGCCUGUGUGUCUAACGUUUUGUUGAUUGAUGGUUAACCAGAAAAUCAAUAUCACUAUGCUAAUGAGACU-GGCAU------ ..((.(((((((((((...)))---------)).))))))))...........((..((((((((..(((....)))...))))))))..)).((((((.......)-)))))------ ( -29.30, z-score = -1.69, R) >droSim1.chr2L 13120360 103 - 22036055 UUGGAGCCUCAAUUGCAUAGCA---------AUCUGGGGCCUGUGUGUCUAACGUUUUGUUGAUUGAUGGUUAACCAGAAAAUCAAUAUCACUAUGCUAAUGAGACU-GGCAU------ ..((.(((((((((((...)))---------)).))))))))...........((..((((((((..(((....)))...))))))))..)).((((((.......)-)))))------ ( -29.30, z-score = -1.69, R) >droPer1.super_5 4699471 112 - 6813705 UUGAAGCCUCAAUUGCAUAGCAAUCUCAUACAUCUCAUACAUGUGUGAGUGGCGUUUUGUUGAUUGAUGGUUAACCGGAAAAUCAAUAUCACUAUGCUAAUGAGACU-AGCAC------ ....((.((((...(((((((...(((((((((.......)))))))))..))((..((((((((..(((....)))...))))))))..)))))))...)))).))-.....------ ( -35.20, z-score = -3.51, R) >dp4.chr4_group5 199005 112 - 2436548 UUGAAGCCUCAAUUGCAUAGCAAUCUCAUACAUCUCAUACAUGUGUGAGUGGCGUUUUGUUGAUUGAUGGUUAACCGGAAAAUCAAUAUCACUAUGCUAAUGAGACU-AGCAU------ ....((.((((...(((((((...(((((((((.......)))))))))..))((..((((((((..(((....)))...))))))))..)))))))...)))).))-.....------ ( -35.20, z-score = -3.46, R) >droWil1.scaffold_180708 7076870 109 + 12563649 UUGGAGCCUCAAUUGCAUAGCAAUC----UCAUUUUGUACAUAUAUGUAUACCGUUUUGUUGAUUGAUGGUUAACCAGAAAAUCAAUAUCACUAUGCUAAUGAGACUAGGCAU------ .....((((...((((...))))((----(((((.((((((....))))))..((..((((((((..(((....)))...))))))))..))......)))))))..))))..------ ( -28.40, z-score = -2.45, R) >droVir3.scaffold_12723 2362495 99 + 5802038 UUGAAGCCUCAAUUGCAUUGCA---------AUCUCA---UUUUGUGUAUGGCGUUUUGUUGAUUGAUGGUUAA-CAGAAAAUCAAUAUCACUAUGCUAAUGAGACA-AGCAC------ ((((....)))).(((.(((..---------.(((((---((....((((((.(...((((((((..((.....-))...)))))))).).)))))).)))))))))-)))).------ ( -25.40, z-score = -1.68, R) >droGri2.scaffold_15126 32461 107 - 8399593 UUGAAGCCUCAAUUGCAUUGCA---------AUCUAA---UUUUGUGUCUGUCGUUUUGUUGAUUGAUGGUUAACCAGGAAAUCAAUAUUGCAAUGCUAAUGAGAAUGAAAACGAGAAC .......((((...((((((((---------(.....---......(.....)....((((((((..(((....)))...)))))))))))))))))...))))............... ( -25.70, z-score = -1.85, R) >consensus UUGGAGCCUCAAUUGCAUAGCA_________AUCUGGGGCCUGUGUGUCUAACGUUUUGUUGAUUGAUGGUUAACCAGAAAAUCAAUAUCACUAUGCUAAUGAGACU_GGCAU______ .......((((...((((((.......................(((.....)))...((((((((..(((....)))...))))))))...))))))...))))............... (-20.55 = -20.39 + -0.16)
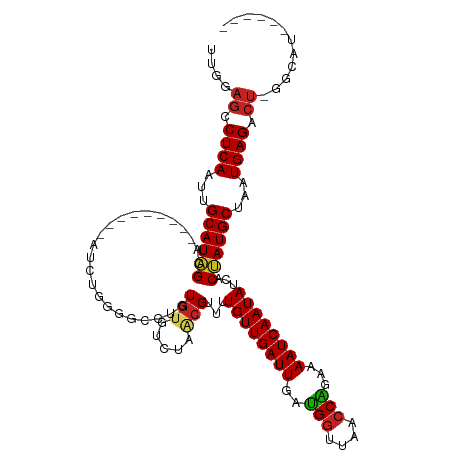
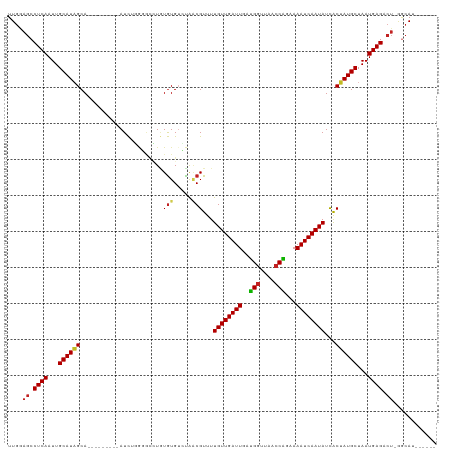
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:37:37 2011