| Sequence ID | dm3.chr2L |
|---|---|
| Location | 1,245,236 – 1,245,327 |
| Length | 91 |
| Max. P | 0.537977 |

| Location | 1,245,236 – 1,245,327 |
|---|---|
| Length | 91 |
| Sequences | 7 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 56.53 |
| Shannon entropy | 0.75982 |
| G+C content | 0.54516 |
| Mean single sequence MFE | -31.03 |
| Consensus MFE | -10.72 |
| Energy contribution | -9.30 |
| Covariance contribution | -1.42 |
| Combinations/Pair | 2.00 |
| Mean z-score | -1.00 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.537977 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 1245236 91 + 23011544 ACUUGAUUCA--CUCGAUUCACUCGCAUUCAGAUGCGUUU-GGACAGAAUUGAGUGAG----UUGCUCGAAGGGCUGCGUUGGG-CCUCGUUCUUGGCG--------------- ....((((((--((((((((.(.(((((....)))))...-)....))))))))))))----))(((.((((((((......))-)))..)))..))).--------------- ( -31.90, z-score = -1.64, R) >droSim1.chr2L 1217420 91 + 22036055 ACUUGAUUCA--CUCGAUUCACUCGCAUUCAGAUGCGUUU-GGACAGAAUUGAGUGAG----UUGCUCGAAGGGCUGCGGUGGG-CCUCGUUCUUGGCG--------------- ....((((((--((((((((.(.(((((....)))))...-)....))))))))))))----))(((.((((((((......))-)))..)))..))).--------------- ( -31.90, z-score = -1.42, R) >droSec1.super_14 1196107 91 + 2068291 ACUUGAUUCA--CUCGAUUCACUCGCAUUCAGAUGCGUUU-GGACAGAAUUGAGUGAG----UUGCUCGAAGGGCUGCGGUGGG-CCUCGUUCUUGGCG--------------- ....((((((--((((((((.(.(((((....)))))...-)....))))))))))))----))(((.((((((((......))-)))..)))..))).--------------- ( -31.90, z-score = -1.42, R) >droYak2.chr2L 1220304 92 + 22324452 AAUCCAUUCA--CUCGAUUCACUCGCAUUCAGAUGCGUUU-GGACAGAAUUGAGUGAG----UUGCUUGAAGGGCUGUGGUGGGAACCCAUUCUUGGGG--------------- ..((((((((--((((((((.(.(((((....)))))...-)....))))))))))))----).(((.....))).......))).((((....)))).--------------- ( -31.70, z-score = -1.88, R) >droEre2.scaffold_4929 1286827 70 + 26641161 ------------CUCGACUCACUCGCAUUCAGAUGCGCUU-GGACAGCAUUGAGUGAG----UUGCUUGAAGGGCCGCGUU----CUUGGG----------------------- ------------..((((((((((((((....))))(((.-....)))...)))))))----)))..(.((((((...)))----))).).----------------------- ( -24.00, z-score = -1.02, R) >droAna3.scaffold_12916 4707160 112 + 16180835 --UCCAUUCAGCUCCAAUCCACUUCAGUUCAGUUGAGUUUUGGACAGGAGCGAGUGAAGCUCUAGCCUGAAGGGUUGUGCGUUGUGUGUGCGCGAGGGCUGUGCGUUUUUACAG --..(((((.(((((..((((.(((((.....)))))...))))..)))))))))).((((((((((.....)))).(((((.......))))))))))).............. ( -35.90, z-score = -0.43, R) >droPer1.super_8 903399 72 + 3966273 -----------UCUGUUUUCUGCCCCGGGCCAUGGGCCAUGGGGGGGGGGGGGGGGGUGCUCCCAUUUGACGAGUCCAGUGUC------------------------------- -----------.(((.(..((.(((((((((...)))).))))).))..)(((((....)))))............)))....------------------------------- ( -29.90, z-score = 0.80, R) >consensus A_UUGAUUCA__CUCGAUUCACUCGCAUUCAGAUGCGUUU_GGACAGAAUUGAGUGAG____UUGCUUGAAGGGCUGCGGUGGG_CCUCGUUCUUGGCG_______________ .................(((((((((((....))))((.....))......)))))))......(((.....)))....................................... (-10.72 = -9.30 + -1.42)

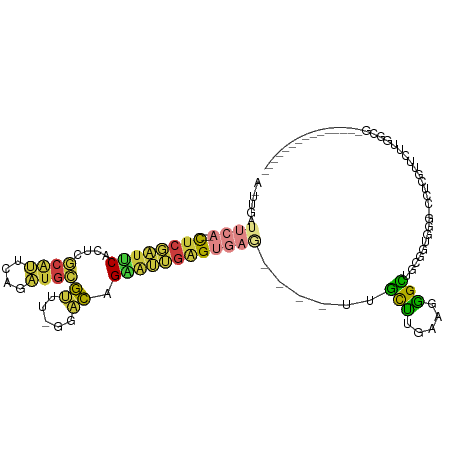

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:08:02 2011