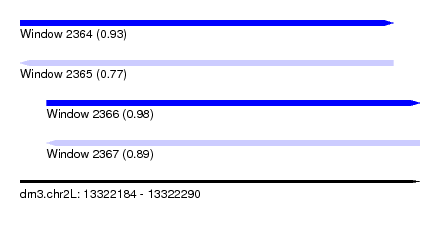
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 13,322,184 – 13,322,290 |
| Length | 106 |
| Max. P | 0.983283 |

| Location | 13,322,184 – 13,322,283 |
|---|---|
| Length | 99 |
| Sequences | 11 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 73.85 |
| Shannon entropy | 0.51296 |
| G+C content | 0.41405 |
| Mean single sequence MFE | -27.53 |
| Consensus MFE | -12.97 |
| Energy contribution | -12.54 |
| Covariance contribution | -0.43 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.929604 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2L 13322184 99 + 23011544 --UCGCUCUGUCUUUUUUCGU-CUGUCUUUUUCGCAACAACAGCGAAAAU---------------UUUCAAUGUUGUACGCGAAAUUGUUGACAAAGUUUGCCUCAAGGCAAACACA --.................((-(...(..((((((.((((((..(((...---------------.)))..))))))..))))))..)..)))...(((((((....)))))))... ( -29.80, z-score = -3.63, R) >droSim1.chr2L 13107955 99 + 22036055 --UCGCUCUGUCUUUUUUCGU-CUGUCUUUUUCGCAACAACAGCGAAAAU---------------UUUCAAUGUUGUACGCGAAAUUGUUGACAAAGUUUGCCUCAAGGCAAACACA --.................((-(...(..((((((.((((((..(((...---------------.)))..))))))..))))))..)..)))...(((((((....)))))))... ( -29.80, z-score = -3.63, R) >droYak2.chr2L 9759376 99 + 22324452 --UAGUUCUGUCUUCUUGCGU-CUGUCUUUUUCGCAACAACAGCGAAAAU---------------UUUCAAUGUUGUACGCGAAAUUGUUGACAAAGUUUGCCUCAAGGCAAACAGA --....(((..........((-(...(..((((((.((((((..(((...---------------.)))..))))))..))))))..)..)))...(((((((....)))))))))) ( -29.90, z-score = -2.98, R) >droEre2.scaffold_4929 14522516 98 - 26641161 --UCGCUCUGUCCUUUUUCGU-CUGUCUUUUUCGCAACAACAGCGAAAUU----------------UUCAAUGUUGUACGCGAAAUUGUUGACAAAGUUUGCCUCAAGGCAAACACA --.................((-(...(..((((((.((((((..((....----------------.))..))))))..))))))..)..)))...(((((((....)))))))... ( -28.90, z-score = -3.51, R) >droAna3.scaffold_12943 1968498 98 + 5039921 --CUAUAUGGUCCUUCCUCGCGCUAUCUUUUUCGCAACAACUGCGAAAAU---------------UUUCAAUGUUGUACGCGAAAUUGUUGACAAAGUUCGCCUCAAGGCAAACA-- --.......(((.....(((((.(((..((((((((.....)))))))).---------------..........)))))))).......)))...(((.(((....))).))).-- ( -21.72, z-score = -1.43, R) >dp4.chr4_group5 182133 91 + 2436548 -----------UCGCGCGCGCAGCCUGUUUUUCGCAACAACAGCAGAAAU---------------UUUCAAUGUUGUACGCGAAAUUGUUGACAAAGUUUGCCUCAAGGCAAACAAA -----------...((((..((((.((..((((((.......)).)))).---------------...))..))))..))))..............(((((((....)))))))... ( -26.00, z-score = -1.16, R) >droPer1.super_5 4682896 91 + 6813705 -----------UCGUGCGCGCAGCCUGUUUUUCGCAACAACAGCAGAAAU---------------UUUCAAUGUUGUACGCGAAAUUGUUGACAAAGUUUGCCUCAAGGCAAACAAA -----------........(((((.....((((((.((((((...((...---------------..))..))))))..))))))..)))).)...(((((((....)))))))... ( -25.60, z-score = -1.16, R) >droWil1.scaffold_180708 7048010 115 - 12563649 --UUAUAUAGUUUUUUUUCUUCUUGUCUUUUUUGCAACAACAGCAAAAAGCAAAAAAAAAAUAAAUUUCAUUGCUGUAUGCGAAAUUGUUGACAAAGUUUGCCUCAAGGCAAACAAA --....................(((((..(((((((...(((((((........................))))))).))))))).....))))).(((((((....)))))))... ( -27.36, z-score = -2.44, R) >droVir3.scaffold_12723 2344646 97 - 5802038 UCUCUUUGGUAUUUGGUCC-UGUGUCCUUUUUCGCAA---CAGCAGCAAU---------------UUUCAAUGUUGUGCGCGGAAUUGUUGACAAAGUUCGCCUCAAGGCAAACAA- ...............(..(-(.(((((..((((((.(---(((((.....---------------......))))))..))))))..)..)))).))..)(((....)))......- ( -23.50, z-score = -0.47, R) >droMoj3.scaffold_6500 6310871 98 + 32352404 UCGCUUUGCCGUUUGGGGCGUGUGUCCUUUUUCGCAA---GAGCAGCAGU---------------UUUCAGUGCAGUGCACGGAACUGUUGACAAAGUUCGCCUCGAGGCCAACAA- .......(((..((((((((..((((....(((....---))).((((((---------------((...((((...)))).)))))))))))).....)))))))))))......- ( -34.60, z-score = -1.15, R) >droGri2.scaffold_15126 14407 94 + 8399593 ----UUUGUGCUUUGGUCC-UGUGUCCUUUUUUGCAAA--CAGCAGCAAU---------------UUUCAAUGCUGUGCGCGGAAUUGUUGACAAAGUUCGCCUCAAGGCAAACAA- ----....((((((((..(-(.(((((..((((((..(--(((((.....---------------......))))))..))))))..)..)))).))..)....))))))).....- ( -25.60, z-score = -0.45, R) >consensus __UCGUUCUGUUUUUGUUCGU_CUGUCUUUUUCGCAACAACAGCAAAAAU_______________UUUCAAUGUUGUACGCGAAAUUGUUGACAAAGUUUGCCUCAAGGCAAACAAA .............................((((((.....(((((..........................)))))...))))))...........(((.(((....))).)))... (-12.97 = -12.54 + -0.43)
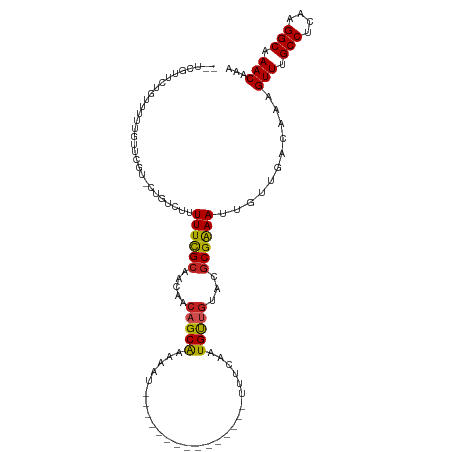
| Location | 13,322,184 – 13,322,283 |
|---|---|
| Length | 99 |
| Sequences | 11 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 73.85 |
| Shannon entropy | 0.51296 |
| G+C content | 0.41405 |
| Mean single sequence MFE | -27.88 |
| Consensus MFE | -12.46 |
| Energy contribution | -11.98 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.767026 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2L 13322184 99 - 23011544 UGUGUUUGCCUUGAGGCAAACUUUGUCAACAAUUUCGCGUACAACAUUGAAA---------------AUUUUCGCUGUUGUUGCGAAAAAGACAG-ACGAAAAAAGACAGAGCGA-- ((((((((((....)))))))((((((.....(((((((.((((((.((((.---------------...)))).)))))))))))))......)-))))).....)))......-- ( -31.60, z-score = -3.04, R) >droSim1.chr2L 13107955 99 - 22036055 UGUGUUUGCCUUGAGGCAAACUUUGUCAACAAUUUCGCGUACAACAUUGAAA---------------AUUUUCGCUGUUGUUGCGAAAAAGACAG-ACGAAAAAAGACAGAGCGA-- ((((((((((....)))))))((((((.....(((((((.((((((.((((.---------------...)))).)))))))))))))......)-))))).....)))......-- ( -31.60, z-score = -3.04, R) >droYak2.chr2L 9759376 99 - 22324452 UCUGUUUGCCUUGAGGCAAACUUUGUCAACAAUUUCGCGUACAACAUUGAAA---------------AUUUUCGCUGUUGUUGCGAAAAAGACAG-ACGCAAGAAGACAGAACUA-- ...(((((((....)))))))((((((.....(((((((.((((((.((((.---------------...)))).))))))))))))).......-.(....)..))))))....-- ( -34.30, z-score = -4.30, R) >droEre2.scaffold_4929 14522516 98 + 26641161 UGUGUUUGCCUUGAGGCAAACUUUGUCAACAAUUUCGCGUACAACAUUGAA----------------AAUUUCGCUGUUGUUGCGAAAAAGACAG-ACGAAAAAGGACAGAGCGA-- ((((((((((....)))))))((((((.....(((((((.((((((.(((.----------------....))).)))))))))))))......)-))))).....)))......-- ( -30.70, z-score = -2.62, R) >droAna3.scaffold_12943 1968498 98 - 5039921 --UGUUUGCCUUGAGGCGAACUUUGUCAACAAUUUCGCGUACAACAUUGAAA---------------AUUUUCGCAGUUGUUGCGAAAAAGAUAGCGCGAGGAAGGACCAUAUAG-- --.(((((((....)))))))...(((.....((((((((.((....))...---------------.(((((((((...))))))))).....))))))))...))).......-- ( -28.80, z-score = -2.43, R) >dp4.chr4_group5 182133 91 - 2436548 UUUGUUUGCCUUGAGGCAAACUUUGUCAACAAUUUCGCGUACAACAUUGAAA---------------AUUUCUGCUGUUGUUGCGAAAAACAGGCUGCGCGCGCGA----------- ...(((((((....)))))))...(((.....(((((((.((((((..((..---------------...))...)))))))))))))....)))(((....))).----------- ( -26.50, z-score = -0.77, R) >droPer1.super_5 4682896 91 - 6813705 UUUGUUUGCCUUGAGGCAAACUUUGUCAACAAUUUCGCGUACAACAUUGAAA---------------AUUUCUGCUGUUGUUGCGAAAAACAGGCUGCGCGCACGA----------- ...(((((((....)))))))...(((.....(((((((.((((((..((..---------------...))...)))))))))))))....)))...........----------- ( -24.90, z-score = -0.66, R) >droWil1.scaffold_180708 7048010 115 + 12563649 UUUGUUUGCCUUGAGGCAAACUUUGUCAACAAUUUCGCAUACAGCAAUGAAAUUUAUUUUUUUUUUGCUUUUUGCUGUUGUUGCAAAAAAGACAAGAAGAAAAAAAACUAUAUAA-- ...(((((((....)))))))((((.(((((((...(((...(((((.((((.......)))).)))))...))).)))))))))))............................-- ( -28.80, z-score = -2.32, R) >droVir3.scaffold_12723 2344646 97 + 5802038 -UUGUUUGCCUUGAGGCGAACUUUGUCAACAAUUCCGCGCACAACAUUGAAA---------------AUUGCUGCUG---UUGCGAAAAAGGACACA-GGACCAAAUACCAAAGAGA -..(((((((....)))))))..((((........((((((((.((.(....---------------).)).)).))---.))))......))))..-((........))....... ( -20.24, z-score = 0.04, R) >droMoj3.scaffold_6500 6310871 98 - 32352404 -UUGUUGGCCUCGAGGCGAACUUUGUCAACAGUUCCGUGCACUGCACUGAAA---------------ACUGCUGCUC---UUGCGAAAAAGGACACACGCCCCAAACGGCAAAGCGA -..(((.(((....))).)))..((((..((((((.((((...)))).))..---------------)))).(((..---..)))......))))...(((......)))....... ( -25.40, z-score = 0.50, R) >droGri2.scaffold_15126 14407 94 - 8399593 -UUGUUUGCCUUGAGGCGAACUUUGUCAACAAUUCCGCGCACAGCAUUGAAA---------------AUUGCUGCUG--UUUGCAAAAAAGGACACA-GGACCAAAGCACAAA---- -..(((((((....))))))).((((.......(((((((((((((.(....---------------).))))).))--)..))......(....).-))).......)))).---- ( -23.84, z-score = -0.36, R) >consensus UUUGUUUGCCUUGAGGCAAACUUUGUCAACAAUUUCGCGUACAACAUUGAAA_______________AUUUUUGCUGUUGUUGCGAAAAAGACAG_ACGAAAAAAAACAGAACGA__ ...(((((((....)))))))...................................................(((.......)))................................ (-12.46 = -11.98 + -0.48)

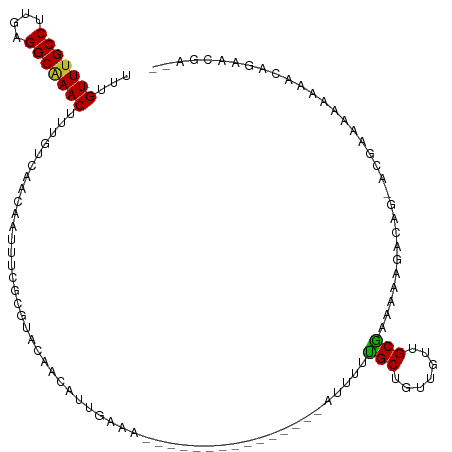
| Location | 13,322,191 – 13,322,290 |
|---|---|
| Length | 99 |
| Sequences | 11 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 75.29 |
| Shannon entropy | 0.48678 |
| G+C content | 0.42178 |
| Mean single sequence MFE | -29.27 |
| Consensus MFE | -14.26 |
| Energy contribution | -13.94 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.13 |
| SVM RNA-class probability | 0.983283 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 13322191 99 + 23011544 --GUCUUUUUUCGU-CUGUCUUUUUCGCAACAACAGCGAAAAU---------------UUUCAAUGUUGUACGCGAAAUUGUUGACAAAGUUUGCCUCAAGGCAAACACAAUGCCAA --..........((-(...(..((((((.((((((..(((...---------------.)))..))))))..))))))..)..)))...(((((((....))))))).......... ( -29.80, z-score = -3.66, R) >droSim1.chr2L 13107962 99 + 22036055 --GUCUUUUUUCGU-CUGUCUUUUUCGCAACAACAGCGAAAAU---------------UUUCAAUGUUGUACGCGAAAUUGUUGACAAAGUUUGCCUCAAGGCAAACACAAUGCCAA --..........((-(...(..((((((.((((((..(((...---------------.)))..))))))..))))))..)..)))...(((((((....))))))).......... ( -29.80, z-score = -3.66, R) >droYak2.chr2L 9759383 99 + 22324452 --GUCUUCUUGCGU-CUGUCUUUUUCGCAACAACAGCGAAAAU---------------UUUCAAUGUUGUACGCGAAAUUGUUGACAAAGUUUGCCUCAAGGCAAACAGAAUGCCAA --((.((((...((-(...(..((((((.((((((..(((...---------------.)))..))))))..))))))..)..)))...(((((((....))))))))))).))... ( -32.10, z-score = -3.69, R) >droEre2.scaffold_4929 14522523 98 - 26641161 --GUCCUUUUUCGU-CUGUCUUUUUCGCAACAACAGCGAAA-U---------------UUUCAAUGUUGUACGCGAAAUUGUUGACAAAGUUUGCCUCAAGGCAAACACAAUGCCAA --..........((-(...(..((((((.((((((..((..-.---------------..))..))))))..))))))..)..)))...(((((((....))))))).......... ( -28.90, z-score = -3.50, R) >droAna3.scaffold_12943 1968505 98 + 5039921 --GUCCUUCCUCGCGCUAUCUUUUUCGCAACAACUGCGAAAAU---------------UUUCAAUGUUGUACGCGAAAUUGUUGACAAAGUUCGCCUCAAGGCAAACA--AUGGCAA --............((((((..((((((.(((((...(((...---------------.)))...)))))..))))))..)........(((.(((....))).))).--))))).. ( -23.30, z-score = -1.46, R) >dp4.chr4_group5 182140 91 + 2436548 -----------CGCAGCCUGUUUUUCGCAACAACAGCAGAAAU---------------UUUCAAUGUUGUACGCGAAAUUGUUGACAAAGUUUGCCUCAAGGCAAACAAAGCGAAGA -----------(((....((((((((((.((((((...((...---------------..))..))))))..)))))).....))))..(((((((....)))))))...))).... ( -27.70, z-score = -2.46, R) >droPer1.super_5 4682903 91 + 6813705 -----------CGCAGCCUGUUUUUCGCAACAACAGCAGAAAU---------------UUUCAAUGUUGUACGCGAAAUUGUUGACAAAGUUUGCCUCAAGGCAAACAAAGCGAAGA -----------(((....((((((((((.((((((...((...---------------..))..))))))..)))))).....))))..(((((((....)))))))...))).... ( -27.70, z-score = -2.46, R) >droWil1.scaffold_180708 7048017 115 - 12563649 --GUUUUUUUUCUUCUUGUCUUUUUUGCAACAACAGCAAAAAGCAAAAAAAAAAUAAAUUUCAUUGCUGUAUGCGAAAUUGUUGACAAAGUUUGCCUCAAGGCAAACAAAAAACUGA --(((((((......(((((..(((((((...(((((((........................))))))).))))))).....))))).(((((((....))))))))))))))... ( -30.56, z-score = -3.05, R) >droVir3.scaffold_12723 2344653 97 - 5802038 GGUAUUU-GGUCCUGUGUCCUUUUUCGCAA--C-AGCAGCAAU---------------UUUCAAUGUUGUGCGCGGAAUUGUUGACAAAGUUCGCCUCAAGGCAAACA-ACUGCCGG ((((.((-((..((.(((((..((((((.(--(-((((.....---------------......))))))..))))))..)..)))).))..)(((....)))...))-).)))).. ( -30.50, z-score = -1.89, R) >droMoj3.scaffold_6500 6310878 97 + 32352404 GCCGUUUGGGGCGUGUGUCCUUUUUCGCAA--G-AGCAGCAGU---------------UUUCAGUGCAGUGCACGGAACUGUUGACAAAGUUCGCCUCGAGGCCAACA-AGAGCCG- .....((((((((..((((....(((....--)-)).((((((---------------((...((((...)))).)))))))))))).....))))))))(((.....-...))).- ( -34.90, z-score = -1.32, R) >droGri2.scaffold_15126 14410 98 + 8399593 -GUGCUUUGGUCCUGUGUCCUUUUUUGCAA--ACAGCAGCAAU---------------UUUCAAUGCUGUGCGCGGAAUUGUUGACAAAGUUCGCCUCAAGGCAAACA-ACUGCCGG -(.((.((((..((.(((((..((((((..--((((((.....---------------......))))))..))))))..)..)))).))..)(((....)))...))-)..))).. ( -26.70, z-score = -0.04, R) >consensus __GUCUUUUUUCGU_CUGUCUUUUUCGCAACAACAGCAAAAAU_______________UUUCAAUGUUGUACGCGAAAUUGUUGACAAAGUUUGCCUCAAGGCAAACA_AAUGCCGA ......................((((((....((((((..........................))))))..))))))...........(((.(((....))).))).......... (-14.26 = -13.94 + -0.32)
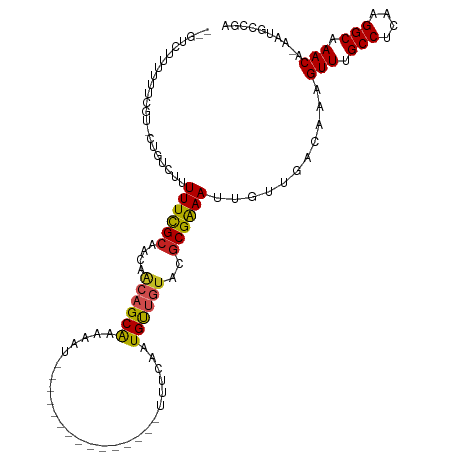
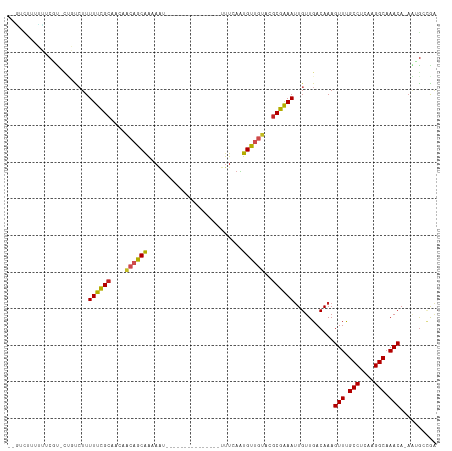
| Location | 13,322,191 – 13,322,290 |
|---|---|
| Length | 99 |
| Sequences | 11 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 75.29 |
| Shannon entropy | 0.48678 |
| G+C content | 0.42178 |
| Mean single sequence MFE | -29.62 |
| Consensus MFE | -12.46 |
| Energy contribution | -11.98 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.893214 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 13322191 99 - 23011544 UUGGCAUUGUGUUUGCCUUGAGGCAAACUUUGUCAACAAUUUCGCGUACAACAUUGAAA---------------AUUUUCGCUGUUGUUGCGAAAAAGACAG-ACGAAAAAAGAC-- ((((((....(((((((....)))))))..))))))...(((((((.((((((.((((.---------------...)))).))))))))))))).......-............-- ( -32.40, z-score = -3.50, R) >droSim1.chr2L 13107962 99 - 22036055 UUGGCAUUGUGUUUGCCUUGAGGCAAACUUUGUCAACAAUUUCGCGUACAACAUUGAAA---------------AUUUUCGCUGUUGUUGCGAAAAAGACAG-ACGAAAAAAGAC-- ((((((....(((((((....)))))))..))))))...(((((((.((((((.((((.---------------...)))).))))))))))))).......-............-- ( -32.40, z-score = -3.50, R) >droYak2.chr2L 9759383 99 - 22324452 UUGGCAUUCUGUUUGCCUUGAGGCAAACUUUGUCAACAAUUUCGCGUACAACAUUGAAA---------------AUUUUCGCUGUUGUUGCGAAAAAGACAG-ACGCAAGAAGAC-- ((((((....(((((((....)))))))..))))))...(((((((.((((((.((((.---------------...)))).))))))))))))).......-.(....).....-- ( -33.60, z-score = -3.64, R) >droEre2.scaffold_4929 14522523 98 + 26641161 UUGGCAUUGUGUUUGCCUUGAGGCAAACUUUGUCAACAAUUUCGCGUACAACAUUGAAA---------------A-UUUCGCUGUUGUUGCGAAAAAGACAG-ACGAAAAAGGAC-- ((((((....(((((((....)))))))..))))))...(((((((.((((((.(((..---------------.-..))).))))))))))))).......-............-- ( -31.50, z-score = -3.09, R) >droAna3.scaffold_12943 1968505 98 - 5039921 UUGCCAU--UGUUUGCCUUGAGGCGAACUUUGUCAACAAUUUCGCGUACAACAUUGAAA---------------AUUUUCGCAGUUGUUGCGAAAAAGAUAGCGCGAGGAAGGAC-- (((.((.--.(((((((....)))))))..)).)))...((((((((.((....))...---------------.(((((((((...))))))))).....))))))))......-- ( -30.30, z-score = -2.29, R) >dp4.chr4_group5 182140 91 - 2436548 UCUUCGCUUUGUUUGCCUUGAGGCAAACUUUGUCAACAAUUUCGCGUACAACAUUGAAA---------------AUUUCUGCUGUUGUUGCGAAAAACAGGCUGCG----------- ....(((...(((((((....)))))))...(((.....(((((((.((((((..((..---------------...))...)))))))))))))....))).)))----------- ( -27.40, z-score = -2.63, R) >droPer1.super_5 4682903 91 - 6813705 UCUUCGCUUUGUUUGCCUUGAGGCAAACUUUGUCAACAAUUUCGCGUACAACAUUGAAA---------------AUUUCUGCUGUUGUUGCGAAAAACAGGCUGCG----------- ....(((...(((((((....)))))))...(((.....(((((((.((((((..((..---------------...))...)))))))))))))....))).)))----------- ( -27.40, z-score = -2.63, R) >droWil1.scaffold_180708 7048017 115 + 12563649 UCAGUUUUUUGUUUGCCUUGAGGCAAACUUUGUCAACAAUUUCGCAUACAGCAAUGAAAUUUAUUUUUUUUUUGCUUUUUGCUGUUGUUGCAAAAAAGACAAGAAGAAAAAAAAC-- ...((((((((((((((....))))))).(((.(((((((...(((...(((((.((((.......)))).)))))...))).)))))))))))))))))...............-- ( -32.60, z-score = -2.81, R) >droVir3.scaffold_12723 2344653 97 + 5802038 CCGGCAGU-UGUUUGCCUUGAGGCGAACUUUGUCAACAAUUCCGCGCACAACAUUGAAA---------------AUUGCUGCU-G--UUGCGAAAAAGGACACAGGACC-AAAUACC (((((((.-.(((((((....))))))).)))))......(((.((((((.((......---------------.....)).)-)--.)))).....)))....))...-....... ( -24.20, z-score = -0.37, R) >droMoj3.scaffold_6500 6310878 97 - 32352404 -CGGCUCU-UGUUGGCCUCGAGGCGAACUUUGUCAACAGUUCCGUGCACUGCACUGAAA---------------ACUGCUGCU-C--UUGCGAAAAAGGACACACGCCCCAAACGGC -.(((...-.(((.(((....))).)))..((((..((((((.((((...)))).))..---------------)))).(((.-.--..)))......))))...)))......... ( -25.90, z-score = 0.38, R) >droGri2.scaffold_15126 14410 98 - 8399593 CCGGCAGU-UGUUUGCCUUGAGGCGAACUUUGUCAACAAUUCCGCGCACAGCAUUGAAA---------------AUUGCUGCUGU--UUGCAAAAAAGGACACAGGACCAAAGCAC- ..(((((.-.(((((((....))))))).)))))......(((((((((((((.(....---------------).))))).)))--..))......(....).))).........- ( -28.10, z-score = -0.68, R) >consensus UCGGCAUU_UGUUUGCCUUGAGGCAAACUUUGUCAACAAUUUCGCGUACAACAUUGAAA_______________AUUUUUGCUGUUGUUGCGAAAAAGACAG_ACGAAAAAAGAC__ ..........(((((((....)))))))...................................................(((.......)))......................... (-12.46 = -11.98 + -0.48)

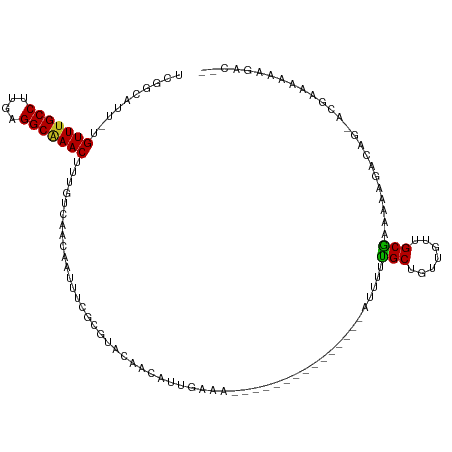
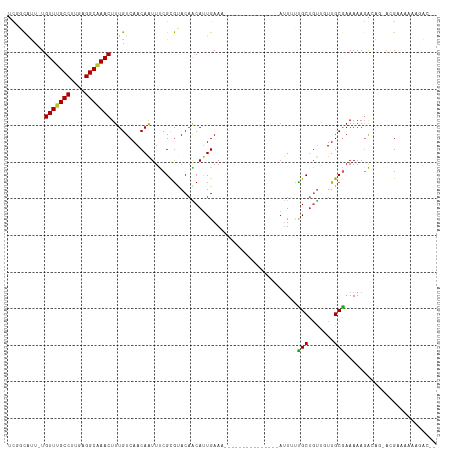
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:37:34 2011