| Sequence ID | dm3.chr2L |
|---|---|
| Location | 13,244,895 – 13,244,987 |
| Length | 92 |
| Max. P | 0.998800 |

| Location | 13,244,895 – 13,244,987 |
|---|---|
| Length | 92 |
| Sequences | 13 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 73.19 |
| Shannon entropy | 0.60946 |
| G+C content | 0.40618 |
| Mean single sequence MFE | -26.82 |
| Consensus MFE | -21.53 |
| Energy contribution | -21.22 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.60 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.50 |
| SVM RNA-class probability | 0.998800 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 13244895 92 + 23011544 AAGAUUUCCGUGGAGAGGAACACUCUAAUGAGUCUAAAAU-A--AUCUUUU-GUAGUGCCCGGCGACUUCGGUAGCUGGGCCA--UAAGGAAUUCAUU-- ..((.((((.((.((((((..((((....)))).......-.--.))))))-.))(.(((((((.((....)).)))))))).--...)))).))...-- ( -30.60, z-score = -2.89, R) >droSim1.chr2L 13028863 93 + 22036055 AAGAUUUCCGUGGAGAGGAACACUCUAAUGAGUCUAAAAU-A--AUUUUUUUGUAGUGCCCGGCGACUUCGGUAGCUGGGCCA--UAAUGAAUUCAUU-- .((((((...(((((.......)))))..)))))).....-(--((((..((((.(.(((((((.((....)).)))))))))--))).)))))....-- ( -28.10, z-score = -2.60, R) >droSec1.super_16 1404345 93 + 1878335 AAGAUUUCCGUGGAGAGGAACACUCUAAUGAGUCUAAAAU-A--AUUUUUUUGUAGUGCCCGGCGACUUUUGUAGCUGGGCCA--UAAGGAAUUCAUU-- .((((((...(((((.......)))))..)))))).....-.--..((((((((.(.(((((((.((....)).)))))))))--)))))))......-- ( -27.60, z-score = -2.51, R) >droYak2.chr2L 9681620 92 + 22324452 AAGAUUUCCGUGGAGAGGAACACUCUAAUGAGUCUAAAAU-A--AUUUUUU-GUAGUGCCCGGUGACUUCGGUUGCUGGGCCA--UAUGGAAUACAUU-- .((((((...(((((.......)))))..)))))).....-.--...(((.-.((..((((((..((....))..))))))..--))..)))......-- ( -31.80, z-score = -3.74, R) >droAna3.scaffold_12943 1889147 94 + 5039921 AAGAUUUCCGUGGAGAGGAGCACUCUAAUGAGUCUAUACU-A--AUUUUUU-GUAGGGCCUAGCAUGACUUCUUGCCGGGCCCGAUAAAUAAUAAAUU-- .((((((...(((((.......)))))..)))))).....-.--(((.(((-((.(((((..(((........)))..))))).))))).))).....-- ( -28.40, z-score = -2.50, R) >dp4.chr4_group4 4127681 97 + 6586962 AAGAUUUCCGUGGAGAGGGGCACUCUAAUGAGUCUAAAAUGA--AUUUUUUCGUAGUGCCCGGCGGCUUAGGUUGCUGGGCCA-AUGUAUAAUACAUUUA .((((((...(((((.......)))))..)))))).(((((.--(((....(((.(.((((((((((....))))))))))).-)))...))).))))). ( -31.60, z-score = -2.35, R) >droPer1.super_10 302741 95 - 3432795 AAGAUUUCCGUGGAGAGGGGCACUCUAAUGAGUCUAAACUGA--AUUUUUUCGUAGUUCCUGGCGACUUCGGUUGCCGGGACA-AUCAAGAAUGAAUG-- .((((((...(((((.......)))))..)))))).......--((((.(((.....((((((((((....))))))))))..-.....))).)))).-- ( -30.60, z-score = -2.24, R) >droEre2.scaffold_4845 4510175 94 + 22589142 AAGAUUUCCGUGGAGAGGAACACUCUAAUGAGUCUAAACUCA--AUUUUUAUUA-AGGCCUGACAGCUUAUGUUAUCGGGCCC-AUUAAUUAUACAUU-- .........(((.((((.....))))..(((((....)))))--..........-.(((((((.(((....))).))))))).-........)))...-- ( -23.50, z-score = -2.31, R) >droWil1.scaffold_180764 2278145 90 + 3949147 AAGAUUUCCGUGGAGAGGUGCACUCUAAUGAGUCUAUUUAGA--AUUUUUCACU-GUGCCUGACUAC-----CAGUCGGGCAA-AUAAUUAAUAAAUAU- .........(((((((((((.((((....)))).))).....--.)))))))).-.((((((((...-----..)))))))).-...............- ( -23.00, z-score = -2.18, R) >droMoj3.scaffold_6500 15268610 94 + 32352404 AAGAUUUCCGUGGAGAGGAGCACUCUAAUGAGUCUAAAUUGA--AUUUUUUGAUUGUGUCUGACGGCGUUUGCUGUCAGGCC--AUAAAGAAUACAUC-- .((((((...(((((.......)))))..))))))....((.--((((((((..((.((((((((((....)))))))))))--))))))))).))..-- ( -29.50, z-score = -3.02, R) >droVir3.scaffold_12723 2252424 92 - 5802038 AAGAUUUCCGUGGAGAGGAACACUCUAAUGAGUCUAA-UGAA--AUUUUUUGCA-GUGCCUGGCCGCGUUUGUUGCCAGGCC--AUAAUUAAUAUGUU-- .((((((...(((((.......)))))..))))))..-....--..........-(.(((((((.((....)).))))))))--..............-- ( -24.40, z-score = -1.49, R) >droGri2.scaffold_15126 7044398 79 + 8399593 AAGAUUUCCGUGGAGAGGAGCGCUCUAAUGAGUCUAAACUCA--AUUUUUUAA--GCGCCUGGCGGCAUUUGCAGCCGGGCC--A--------------- .((((((...(((((.......)))))..)))))).......--.........--(.(((((((.((....)).))))))))--.--------------- ( -26.20, z-score = -0.96, R) >anoGam1.chr3L 19691472 95 - 41284009 AAGAUUUCCGUGGAGAGACAUAUUCUAAUGAGACUAAACUGAUCAUUUAUCAUUUGAUCUUGAU-ACUAUCGUGCUUAUAGUA--UUUUUAUUUCAAU-- ....((((....))))............(((((.((((..(((((.........)))))..(((-(((((.......))))))--)))))))))))..-- ( -13.40, z-score = 0.67, R) >consensus AAGAUUUCCGUGGAGAGGAACACUCUAAUGAGUCUAAAAU_A__AUUUUUU_GUAGUGCCUGGCGACUUUUGUUGCCGGGCCA_AUAAAGAAUACAUU__ .((((((...(((((.......)))))..))))))......................(((((((.((....)).)))))))................... (-21.53 = -21.22 + -0.31)

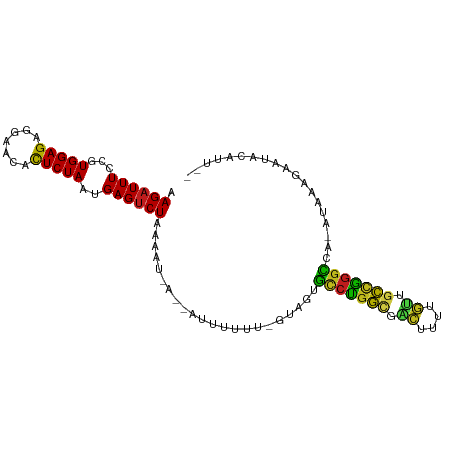
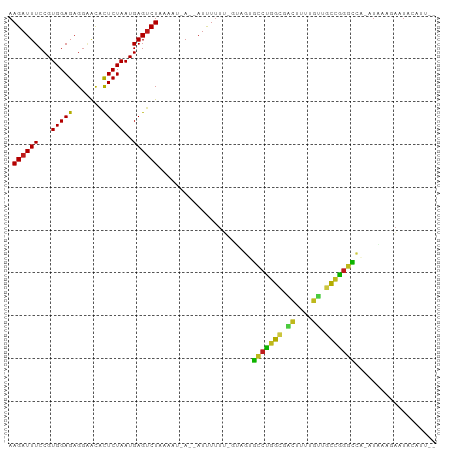
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:37:28 2011