| Sequence ID | dm3.chr2L |
|---|---|
| Location | 13,209,552 – 13,209,643 |
| Length | 91 |
| Max. P | 0.850116 |

| Location | 13,209,552 – 13,209,643 |
|---|---|
| Length | 91 |
| Sequences | 10 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 61.22 |
| Shannon entropy | 0.77913 |
| G+C content | 0.34716 |
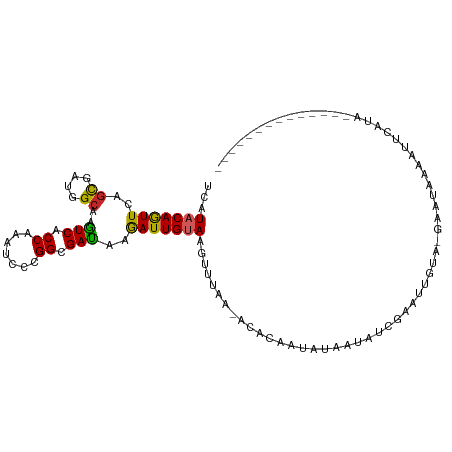
| Mean single sequence MFE | -15.51 |
| Consensus MFE | -8.41 |
| Energy contribution | -7.65 |
| Covariance contribution | -0.76 |
| Combinations/Pair | 1.36 |
| Mean z-score | -0.35 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.850116 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
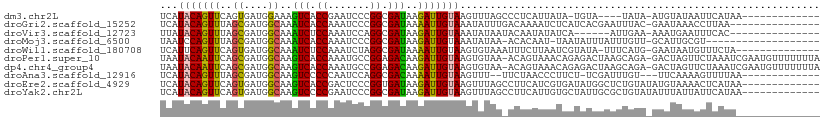
>dm3.chr2L 13209552 91 + 23011544 UCAUACAGUUCAGUGAUGGAAAGUCACCGAAUCCCGGCGAUAAGAUUGUAAGUUUAGCCCUCAUUAUA-UGUA----UAUA-AUGUAUAAUUCAUAA------------- ...(((((((((....))....(((.(((.....))).)))..)))))))...........(((((((-....----))))-)))............------------- ( -14.80, z-score = -0.39, R) >droGri2.scaffold_15252 4049239 94 + 17193109 UCAUACAGUUUAGCGAUGGCAAAUCACCAAAUCCCGGCGAUAAAAUUGUAAAUAUUUGACAAAAUCUCAUCACGAAUUUAC-GAAUAAACCUUAA--------------- ...((((((((.((....))..(((.((.......)).))).))))))))..((((((..(((.((.......)).))).)-)))))........--------------- ( -12.10, z-score = -0.56, R) >droVir3.scaffold_12723 2219226 88 - 5802038 UUAUACAGUUUAGCGAUGGCAAAUCUCCAAAUCCAGGCGAUAAGAUUGUAAAUAUAAUACAAUAUAUCA------AUUGAA-AAAUGAAUUUCAC--------------- ...((((((((.((....))..(((.((.......)).))).))))))))...................------..((((-(......))))).--------------- ( -11.60, z-score = -0.58, R) >droMoj3.scaffold_6500 15231309 88 + 32352404 UAAUCCAGUUUAGCGAUGGCAAAUCACCAAAUCCCGGCGAUAAGAUUGUAAAUAUAA-ACACAAU-UAAUAUUUAUUUGUU-GCAUUGCGU------------------- ............((((((.((((((.((.......)).)))..((((((........-..)))))-)............))-)))))))..------------------- ( -14.90, z-score = -0.56, R) >droWil1.scaffold_180708 403223 94 - 12563649 UCAUUCAGUUCAGUGAUGGCAAAUCUCCAAAUCUAGGCGAUAAAAUUGUAAGUGUAAAUUUCUUAAUCGUAUA-UUUCAUG-GAAUAAUGUUUCUA-------------- .((((..((((.((((..((..(((.((.......)).))).......((((.........))))...))...-..)))).-))))))))......-------------- ( -9.90, z-score = 0.94, R) >droPer1.super_10 260249 108 - 3432795 UAAUACAAUUCAGCGAUGGCAAGUCACCAAAUGCCGGAGACAAGAUUGUAAGUGUAA-ACAGUAAACAGAGACUAAGCAGA-GACUAGUUCUAAAUCGAAUGUUUUUUUA ...(((((((..((..(((((..........)))))..).)..))))))).....((-(((........((((((......-...))).)))........)))))..... ( -15.79, z-score = 0.70, R) >dp4.chr4_group4 2456623 108 - 6586962 UAAUACAAUUCAGCGAUGGCAAGUCACCAAAUGCCGGAGACAAGAUUGUAAGUGUAA-ACAGUAAACAGAGACUAAGCAGA-GACUAGUUCUAAAUCGAAUGUUUUUUUA ...(((((((..((..(((((..........)))))..).)..))))))).....((-(((........((((((......-...))).)))........)))))..... ( -15.79, z-score = 0.70, R) >droAna3.scaffold_12916 10858351 91 + 16180835 UCAUACAGUUUAGCGAUGGCAAGUCCCCCAAUCCAGGCGACAAAAUUGUAAGUUU--UUCUAACCCUUCU-UCGAUUUGU---UUCAAAAGUUUUAA------------- ...((((((((.((....))..(((((........)).))).)))))))).....--.............-..((.....---.))...........------------- ( -12.00, z-score = -0.07, R) >droEre2.scaffold_4929 14411386 97 - 26641161 UCAUACAGUUCAGUGAUGGCAAGUCACCGACUCCCGGUGAUAAGAUUGUAAGUUUAGCCUUCAUCGUGAUAUGGCUCUGUAUAUGUAAAACUCAUAA------------- ..((((((.(((((((.(((..(((((((.....))))))).((((.....)))).))).))))..))).......))))))...............------------- ( -23.20, z-score = -1.21, R) >droYak2.chr2L 9633264 97 + 22324452 UCAUACAGUUCAGUGAUGGCAAGUCCCCGAAUCCCGGCGAUAAGAUUGUAAGUUUAGCCUUCAUUGUGCUAUUGCGCUGUAUAUUUAUUAUUCAUAA------------- ..(((((((.((((((.(((..(((.(((.....))).))).((((.....)))).))).)))))).((....)))))))))...............------------- ( -25.00, z-score = -2.48, R) >consensus UCAUACAGUUCAGCGAUGGCAAGUCACCAAAUCCCGGCGAUAAGAUUGUAAGUUUAA_ACACAAUAUAAUAUCGAAUUGUA_GAAUAAAAUUCAUA______________ ...(((((((..((....))..(((.((.......)).)))..)))))))............................................................ ( -8.41 = -7.65 + -0.76)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:37:17 2011