
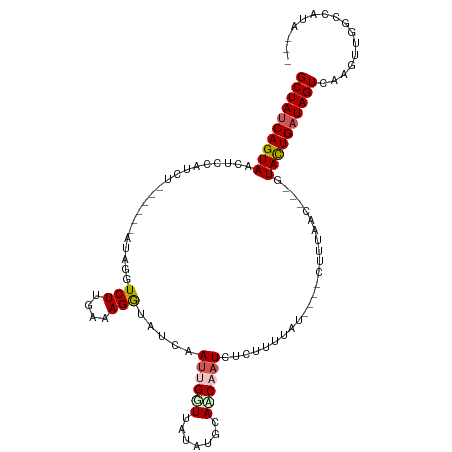
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 13,194,996 – 13,195,094 |
| Length | 98 |
| Max. P | 0.646162 |

| Location | 13,194,996 – 13,195,094 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 67.80 |
| Shannon entropy | 0.54725 |
| G+C content | 0.31563 |
| Mean single sequence MFE | -19.23 |
| Consensus MFE | -9.25 |
| Energy contribution | -9.89 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>dm3.chr2L 13194996 98 + 23011544 GCUAUCAGUAAUUCCAUCU------AUAGGUCUGGAAAGGUUUCAAUUGCUUAUUUGCAACAAUUUCUUUUGU----CUUUAAC---GUACUGAUAGUCUAGUUGGCCAUU--- ((((((((((.(((((...------.......)))))..(((.....(((......)))((((......))))----....)))---.)))))))))).............--- ( -19.10, z-score = -1.13, R) >droSim1.chr2L 12989412 98 + 22036055 GCUAUCAGUAACUCCAUCU------AUAGGUCUUGAAAGGUCUCAAUUGUUUAUUUGCAACAAUCUCUUUUAU----CUUUAAU---GUACUGAUAGUCAAGUUGGCCAUU--- ((((((((((....(((..------..(((...(((((((......((((......)))).....))))))).----)))..))---))))))))))).............--- ( -16.90, z-score = -0.91, R) >droSec1.super_16 1349549 99 + 1878335 GCUAUCAGUAACUCCAUCU------AUAGGUCUUGAAAGGUCUCAAUUGGUUAUAUGCAACAAUCUGUUUUAU----CUUUAACUG-AUACUG-UAGUCAAGUUGAGAGUC--- (((.....(((((....((------((((....(((......)))((..((((.(((.(((.....))).)))----...))))..-)).)))-)))...)))))..))).--- ( -15.80, z-score = 0.76, R) >droYak2.chr2L 9618604 114 + 22324452 GCUAUCAGUAACUCUAUCACAAGAAAUAGCUCUUGAUAGAUAUCAAUUGAUUUCACCAAUCUAUUUUAUCUAUGGGCUAAUAUUAAUGUAUUGAUAGUUAAGAAUGUUUUAAAA ((((((((((..(((......)))..((((((...(((((((..(((.((((.....)))).))).))))))))))))).........))))))))))................ ( -23.50, z-score = -2.12, R) >droEre2.scaffold_4929 14395788 108 - 26641161 GCUAUCAGUAAUUCUAUCACAACAAAUAGCUCUUGAUAGGCAUCAAUUGGUU--GACAAUCUUUCUUUGUUAU----CAAUAGCUUUAUAUUGAUAGUCAAUUUAGUUUUAAAU ((((((((((..(((((((..............))))))).....((((((.--(((((.......)))))))----)))).......))))))))))................ ( -20.84, z-score = -2.20, R) >consensus GCUAUCAGUAACUCCAUCU______AUAGGUCUUGAAAGGUAUCAAUUGGUUAUAUGCAACAAUCUCUUUUAU____CUUUAAC___GUACUGAUAGUCAAGUUGGCCAUA___ ((((((((((...............(((((..(((((....))))).(((......))).........)))))...............))))))))))................ ( -9.25 = -9.89 + 0.64)


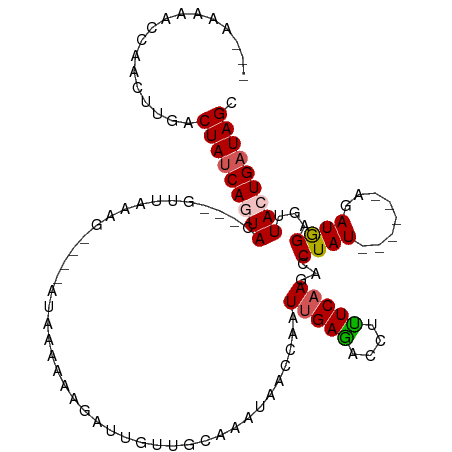
| Location | 13,194,996 – 13,195,094 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 67.80 |
| Shannon entropy | 0.54725 |
| G+C content | 0.31563 |
| Mean single sequence MFE | -17.01 |
| Consensus MFE | -9.98 |
| Energy contribution | -9.98 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.17 |
| Mean z-score | -0.82 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.646162 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 13194996 98 - 23011544 ---AAUGGCCAACUAGACUAUCAGUAC---GUUAAAG----ACAAAAGAAAUUGUUGCAAAUAAGCAAUUGAAACCUUUCCAGACCUAU------AGAUGGAAUUACUGAUAGC ---..............(((((((((.---.......----........(((((((.......))))))).......(((((.......------...))))).))))))))). ( -18.80, z-score = -2.04, R) >droSim1.chr2L 12989412 98 - 22036055 ---AAUGGCCAACUUGACUAUCAGUAC---AUUAAAG----AUAAAAGAGAUUGUUGCAAAUAAACAAUUGAGACCUUUCAAGACCUAU------AGAUGGAGUUACUGAUAGC ---..............((((((((((---......(----(.......(((((((.......)))))))........))....((...------....)).).))))))))). ( -17.46, z-score = -1.31, R) >droSec1.super_16 1349549 99 - 1878335 ---GACUCUCAACUUGACUA-CAGUAU-CAGUUAAAG----AUAAAACAGAUUGUUGCAUAUAACCAAUUGAGACCUUUCAAGACCUAU------AGAUGGAGUUACUGAUAGC ---(((((.((.(((((...-..((.(-(((((....----(((.(((.....)))...)))....)))))).))...)))))..(...------.).)))))))......... ( -12.40, z-score = 1.11, R) >droYak2.chr2L 9618604 114 - 22324452 UUUUAAAACAUUCUUAACUAUCAAUACAUUAAUAUUAGCCCAUAGAUAAAAUAGAUUGGUGAAAUCAAUUGAUAUCUAUCAAGAGCUAUUUCUUGUGAUAGAGUUACUGAUAGC .................((((((..((........(((((.(((((((.(((.((((.....)))).)))..)))))))...).))))..(((......)))))...)))))). ( -17.90, z-score = -0.33, R) >droEre2.scaffold_4929 14395788 108 + 26641161 AUUUAAAACUAAAUUGACUAUCAAUAUAAAGCUAUUG----AUAACAAAGAAAGAUUGUC--AACCAAUUGAUGCCUAUCAAGAGCUAUUUGUUGUGAUAGAAUUACUGAUAGC .................((((((.((.....(((((.----((((((((........(((--((....)))))((((....)).))..)))))))))))))...)).)))))). ( -18.50, z-score = -1.55, R) >consensus ___AAAAACCAACUUGACUAUCAGUAC___GUUAAAG____AUAAAAAAGAUUGUUGCAAAUAACCAAUUGAGACCUUUCAAGACCUAU______AGAUGGAGUUACUGAUAGC .................(((((((((..........................................(((((....)))))...((((........))))...))))))))). ( -9.98 = -9.98 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:37:15 2011