
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 13,194,512 – 13,194,605 |
| Length | 93 |
| Max. P | 0.749772 |

| Location | 13,194,512 – 13,194,605 |
|---|---|
| Length | 93 |
| Sequences | 8 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 65.80 |
| Shannon entropy | 0.67700 |
| G+C content | 0.34958 |
| Mean single sequence MFE | -12.97 |
| Consensus MFE | -6.47 |
| Energy contribution | -5.68 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.40 |
| Mean z-score | -0.72 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.749772 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 13194512 93 + 23011544 AGCCACCAAUGAAUGUGAUUUGAUUUACAAAUAUAUUUAAACAUAUU----UCCAGGUCAUGCAGAUGCACACCUUCAUUCCAGAAGCAUUUACCGU .((....((((((.(((...((((((..((((((........)))))----)..))))))(((....)))))).))))))......))......... ( -15.20, z-score = -1.18, R) >droSim1.chr2L 12988928 93 + 22036055 AGCCACCAAUGAUUGCGAUUUGUUUUACACAUAUAUUUCAUCUUAUU----UGCAGGUCAUGCAGAUGCACACCUUCAUUCCAGAAGCAUUUACCGU .........(((.(((((..(((.........)))..)).(((....----((.((((..(((....))).)))).))....))).))).))).... ( -13.50, z-score = -0.01, R) >droSec1.super_16 1349065 93 + 1878335 AGCCACCACUGAUUGCGAUUUGUUUUACACAUAUAUUUAAUCUUAUU----UGCAGGUCAUGCAGAUGCACACCUUCAUUCCAGAAGCAUUUACCGU .((.......(((((..((.(((.....))).))...)))))..(((----((((.....)))))))))....((((......)))).......... ( -12.60, z-score = 0.24, R) >droYak2.chr2L 9618117 88 + 22324452 ----AUCCGCUUAUCCCAUUU-UAUUACAAACAUAUUUGAUCUUAUU----UGCAGGUUAUGCAGAUGCACACCUUUAUUCCAGAAGCAUUUACCGU ----....((((.........-.....((((....))))....((((----((((.....))))))))................))))......... ( -11.70, z-score = -1.08, R) >droEre2.scaffold_4929 14395292 97 - 26641161 AUCCGCCCAUAAAUACGAUUUAUUUUACAAAUAUUAUUUAUCUUAUUAUUUCUUAGGUUAUGCAGAUGCACACCUUCAUUCCAGAAGCAUUUACCGU ....((..(((((((..((((.......))))..)))))))........((((.((((..(((....))).)))).......))))))......... ( -8.40, z-score = -0.06, R) >dp4.chr4_group4 2437769 85 - 6586962 ------------GUAUUGUGUAUAACAAAAUUUAAUAUAUUCAAUCUUUUAAAUAGGUUAUGCAGAUGCACACUCUCAUCCCGGAGGCAUUUACCGU ------------(((((.((((((((...((((((.............))))))..)))))))))))))...((((......))))........... ( -16.12, z-score = -1.60, R) >droPer1.super_10 241515 85 - 3432795 ------------GUAUUUUGUAUAACAAAAUUUAAUAUAUUCAAUCUUUUAAAUAGGUUAUGCAGAUGCACACUCUCAUCCCGGAGGCAUUUACCGU ------------((((((.(((((((...((((((.............))))))..)))))))))))))...((((......))))........... ( -16.12, z-score = -2.14, R) >droWil1.scaffold_180708 384067 78 - 12563649 ----------------UUUAAAGACCAAAGUUACCUUUUAUCUCUCAUUU---UAGGUCAUGCAGAUGCAUACACUUAUCCCAGAGGCAUUUACUGU ----------------........................(((((.....---(((((.((((....))))..)))))....))))).......... ( -10.10, z-score = 0.05, R) >consensus ____ACC__U_AAUACGAUUUAUAUUACAAAUAUAUUUAAUCUUAUU____UGCAGGUCAUGCAGAUGCACACCUUCAUUCCAGAAGCAUUUACCGU .......................................................(((..(((....)))...((((......)))).....))).. ( -6.47 = -5.68 + -0.80)

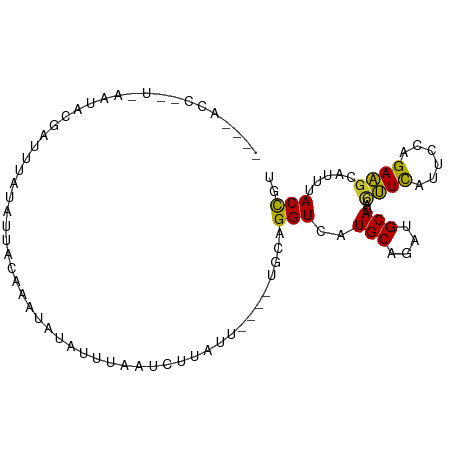

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:37:13 2011