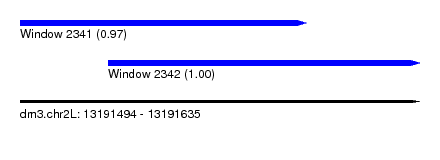
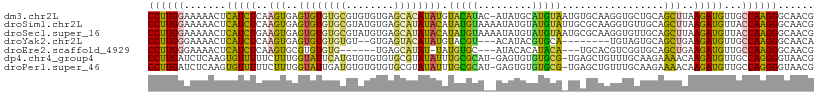
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 13,191,494 – 13,191,635 |
| Length | 141 |
| Max. P | 0.999519 |

| Location | 13,191,494 – 13,191,595 |
|---|---|
| Length | 101 |
| Sequences | 7 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 69.02 |
| Shannon entropy | 0.56557 |
| G+C content | 0.37792 |
| Mean single sequence MFE | -26.24 |
| Consensus MFE | -7.79 |
| Energy contribution | -5.13 |
| Covariance contribution | -2.67 |
| Combinations/Pair | 2.00 |
| Mean z-score | -2.95 |
| Structure conservation index | 0.30 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.83 |
| SVM RNA-class probability | 0.970249 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
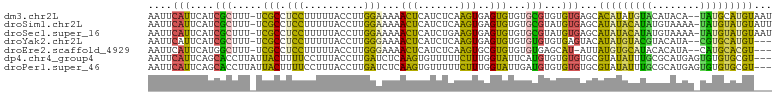
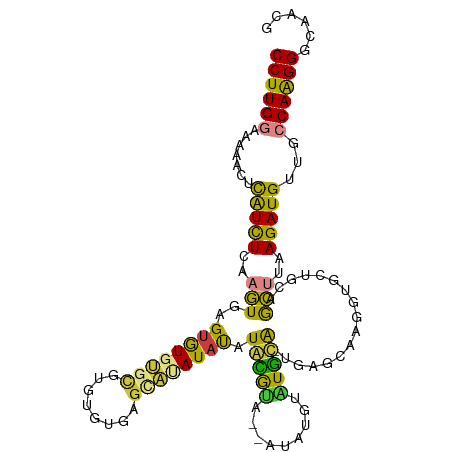
>dm3.chr2L 13191494 101 + 23011544 AAUUCAUUCAUCGCUUU-UCGCCUCCUUUUUACCUUGGAAAAACUCAUCUCAAGUGAGUGUGUGCGUGUGUGAGCACAUAUGUACAUACA--UAUGCAUGUAAU ....(((.((((((((.-........(((((......))))).........))))))((((((((((((((....)))))))))))))).--..)).))).... ( -24.67, z-score = -1.97, R) >droSim1.chr2L 12986039 102 + 22036055 AAUUCAUUCAUCGCUUU-UCGCCUCCUUUUUACCUUGGAAAAACUCAUCUCAAGUGAGUGUGUGCGUAUGUGAGCAUAUACAUAUGUAAAA-UAUGUAUGUAUU ..(((((....(((...-.(((.(((..........)))...((((((.....))))))))).)))...))))).((((((((((.....)-)))))))))... ( -24.60, z-score = -3.13, R) >droSec1.super_16 1346174 102 + 1878335 AAUUCAUUCAUCGCUUU-UCGCCUCCUUUUUACCUUGGAAAAACUCAUCUGAAGUGAGUGUGUGCGUAUGUGAGCAUAUACAUAUGUAAAA-UAUGUAUGUAAU ..(((((....(((...-.(((.(((..........)))...((((((.....))))))))).)))...))))).((((((((((.....)-)))))))))... ( -24.60, z-score = -2.86, R) >droYak2.chr2L 9615113 98 + 22324452 AAUUCAUUCAUCGCUUU-UCGCCUCCUUUUUACCUUGGGAAAACUCAUCUCAAGUGAGUGUGUGUGUGUGAGUACAUAUGUACGUACAUA--CGUGCAUGU--- .((((((.((((((.((-((.((.............))))))((((((.....))))))))).))).))))))..((((((((((....)--)))))))))--- ( -27.62, z-score = -2.65, R) >droEre2.scaffold_4929 14392252 97 - 26641161 AAUUCAUUCAUGGCUUU-UCGCCUCCUUUUUACCUUGGGAAAACUCAUCUCAAGUGCGUGUGUGUGAGCAU-AUUAUGUGCAUACACAUA--CAUGCACGU--- ...........(((...-..)))...........((((((.......))))))(((((((((((((.((((-.....))))...))))))--)))))))..--- ( -32.10, z-score = -4.29, R) >dp4.chr4_group4 913304 101 + 6586962 AAUUCAUUCAGCACCUUAUUACUUUUCCUUUACCUUGAUCUCAAGUGUUUUUCUUUGGUAUUCAUGUGUGUGUGCGUAUAUUUGCGCAUGAGUGUGUGCGU--- .((.(((((.((((.....((((........((((((....)))).))........)))).....))))..(((((((....)))))))))))).))....--- ( -23.79, z-score = -2.53, R) >droPer1.super_46 442621 101 - 590583 AAUUCAUUCAGCACCUUAUUACUUUUCCUUUACCUUGAUCUCAAGUGUUUUUCUUUGGUAUUGAUGUGUGUGUGCGUAUAUUUGCGCAUGAGUGUGUGCGU--- .((.(((((.((((.(((.((((........((((((....)))).))........)))).))).))))..(((((((....)))))))))))).))....--- ( -26.29, z-score = -3.19, R) >consensus AAUUCAUUCAUCGCUUU_UCGCCUCCUUUUUACCUUGGAAAAACUCAUCUCAAGUGAGUGUGUGUGUGUGUGAGCAUAUAUAUACGCAUAA_UAUGUAUGU___ ............((............................(((.......)))..(((((((.(((((....))))).)))))))........))....... ( -7.79 = -5.13 + -2.67)
| Location | 13,191,525 – 13,191,635 |
|---|---|
| Length | 110 |
| Sequences | 7 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 66.09 |
| Shannon entropy | 0.64315 |
| G+C content | 0.44215 |
| Mean single sequence MFE | -36.73 |
| Consensus MFE | -17.73 |
| Energy contribution | -16.73 |
| Covariance contribution | -1.00 |
| Combinations/Pair | 1.69 |
| Mean z-score | -2.89 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.97 |
| SVM RNA-class probability | 0.999519 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
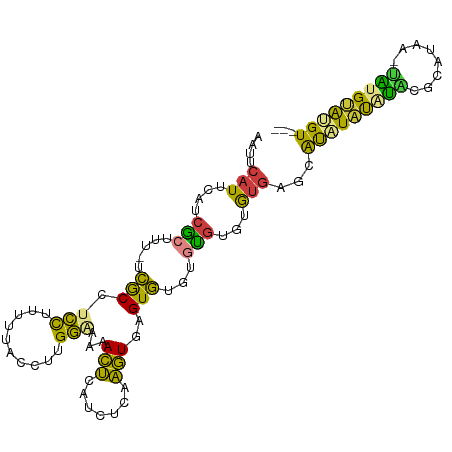
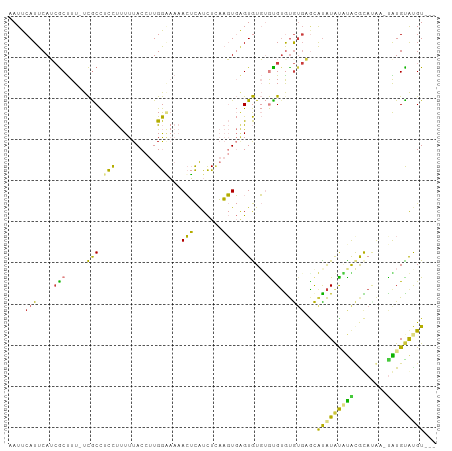
>dm3.chr2L 13191525 110 + 23011544 CCUUGGAAAAACUCAUCUCAAGUGAGUGUGUGCGUGUGUGAGCACAUAUGUACAUAC-AUAUGCAUGUAAUGUGCAAGGUGCUGCAGCUUAAGAUGUUGCCAAGGGCAACG ((((((.......(((((.((((..((((((((((((((....))))))))))))))-...(((((.....)))))..........)))).)))))...))))))...... ( -41.80, z-score = -3.11, R) >droSim1.chr2L 12986070 111 + 22036055 CCUUGGAAAAACUCAUCUCAAGUGAGUGUGUGCGUAUGUGAGCAUAUACAUAUGUAAAAUAUGUAUGUAUUGCGCAAGGUGUUGCAGCUUAAGAUGUUACCAAGGGCAACG ((((((....((((((.....))))))....((((...(((((((((((((((.....)))))))))).....((((....)))).)))))..))))..))))))...... ( -35.90, z-score = -3.09, R) >droSec1.super_16 1346205 111 + 1878335 CCUUGGAAAAACUCAUCUGAAGUGAGUGUGUGCGUAUGUGAGCAUAUACAUAUGUAAAAUAUGUAUGUAAUGCGCAAGGUGUUGCAGCUUAAGAUGUUACCAAGGGCAACG ((((((....((((((.....))))))....((((...(((((..((((((((.....))))))))(((((((.....))))))).)))))..))))..))))))...... ( -36.10, z-score = -3.39, R) >droYak2.chr2L 9615144 98 + 22324452 CCUUGGGAAAACUCAUCUCAAGUGAGUGUGUGUGU--GUGAGUACAUAUGUACGU---ACAUACGUGCA--------UGUAGUGCAGCUGAAGAUGUUGCCAAGGGCAACA ((((((.((....(((((..(((..(((..(((((--(((.((((....)))).)---)))))))..))--------)........)))..))))))).))))))...... ( -36.20, z-score = -2.72, R) >droEre2.scaffold_4929 14392283 98 - 26641161 CCUUGGGAAAACUCAUCUCAAGUGCGUGUGUG------UGAGCAUAU-UAUGUGC---AUACACAUACA---UGCACGUCGGUGCAGCUGAAGAUGUUGCCAAGGGCAACG ((((((.((....(((((...(((((((((((------((.((((..-...))))---...))))))))---))))).((((.....))))))))))).))))))...... ( -41.40, z-score = -3.96, R) >dp4.chr4_group4 913336 109 + 6586962 CCUUGAUCUCAAGUGUUUUUCUUUGGUAUUCAUGUGUGUGUGCGUAUAUUUGCGCAU-GAGUGUGUGCG-UGAGCUGUUUGCAAGAAAACAAGAUGUUGCCAGGGGUAACG .((((....)))).(((..(((((((((.((.(((.....((((.(((((..(((((-......)))))-..)).))).)))).....))).))...))))))))).))). ( -33.70, z-score = -2.18, R) >droPer1.super_46 442653 109 - 590583 CCUUGAUCUCAAGUGUUUUUCUUUGGUAUUGAUGUGUGUGUGCGUAUAUUUGCGCAU-GAGUGUGUGCG-UGAGCUGUUUGCAAGAAAACAAGAUGUUGCCAGGGGUAACG .((((....)))).(((..(((((((((......(((...((((.(((((..(((((-......)))))-..)).))).)))).....)))......))))))))).))). ( -32.00, z-score = -1.81, R) >consensus CCUUGGAAAAACUCAUCUCAAGUGAGUGUGUGCGUGUGUGAGCAUAUAUAUACGUA__AUAUGUAUGCA_UGAGCAAGGUGCUGCAGCUUAAGAUGUUGCCAAGGGCAACG ((((((.......(((((..(((..((((((((........))))))))(((((((.....)))))))..................)))..)))))...))))))...... (-17.73 = -16.73 + -1.00)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:37:12 2011