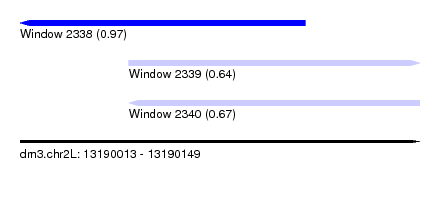
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 13,190,013 – 13,190,149 |
| Length | 136 |
| Max. P | 0.971827 |

| Location | 13,190,013 – 13,190,110 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 86.80 |
| Shannon entropy | 0.22636 |
| G+C content | 0.41081 |
| Mean single sequence MFE | -25.66 |
| Consensus MFE | -19.36 |
| Energy contribution | -20.28 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.971827 |
| Prediction | RNA |
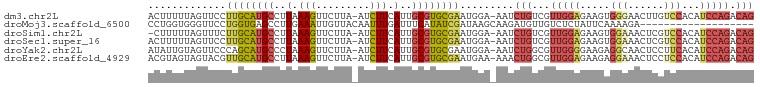
Download alignment: ClustalW | MAF
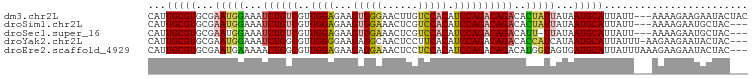
>dm3.chr2L 13190013 97 - 23011544 CAUUGCGUGCGAAUGGAAAUCUGUCGUUGGAGAAGUGGGAACUUGUCCACAUCCAGACAGACACUAUUAUAAUGCAUUAUU---AAAAGAAGAAUACUAC ...(((((...(((((...((((((..((((...(((((......))))).))))))))))..)))))...))))).....---................ ( -26.90, z-score = -3.16, R) >droSim1.chr2L 12984590 94 - 22036055 CAUUGCGUGCGAAUGGAAAUCUGUCGUUGGAGAAGUGGAAACUCGUCCACAUCCAGACAGACACUAUUAUAAUGCAUUAUU---AAAAGAAUGCUAC--- ...(((((...(((((...((((((..((((...(((((......))))).))))))))))..)))))...))))).....---.............--- ( -27.80, z-score = -3.45, R) >droSec1.super_16 1344686 93 - 1878335 CAUUGCGUGCGAAUGGAAAUCUGUCGUUGGAGAAGUGGAAACUCGUCCACAUCCAGACAGACAUU-UUAUAAUGCAUUAUU---AAAAGAAUGCUAC--- ...(((((..(((((....((((((..((((...(((((......))))).))))))))))))))-)....))))).....---.............--- ( -24.90, z-score = -2.42, R) >droYak2.chr2L 9613589 96 - 22324452 CAUUGCGUGCGAAUGGAAAUCUGGCGUUGGGGAAGAGGCAACUCCUUCACAUCCAGACAGACACCAUCAUAAUGCAUUAUUU-AAGAAGAAUACUAC--- ...(((((....((((...((((...(((((((((((....)).))))...))))).))))..))))....)))))......-..............--- ( -24.10, z-score = -1.88, R) >droEre2.scaffold_4929 14390780 97 + 26641161 CAUUGCGUGCGAAUGAAAAACUGGCGUUGGAGAAGAGGAAACUCCUCCACAUCCAGACAGACAUGGUAGUGAUGCAUUAUUUAAAGAAGAAUACUAC--- (((..(.(((..(((.....((((...(((((..(((....))))))))...)))).....))).))))..))).......................--- ( -24.60, z-score = -2.44, R) >consensus CAUUGCGUGCGAAUGGAAAUCUGUCGUUGGAGAAGUGGAAACUCGUCCACAUCCAGACAGACACUAUUAUAAUGCAUUAUU___AAAAGAAUACUAC___ ...(((((...(((((...((((((..((((...(((((......))))).))))))))))..)))))...)))))........................ (-19.36 = -20.28 + 0.92)

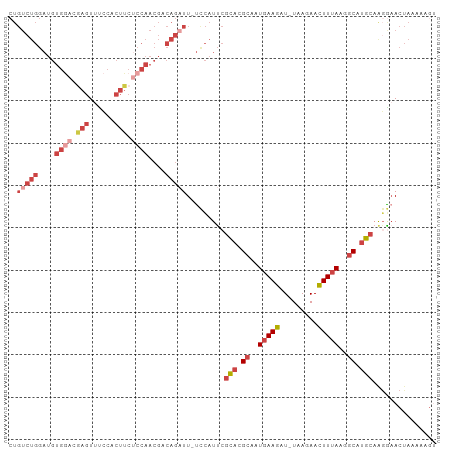
| Location | 13,190,050 – 13,190,149 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 72.91 |
| Shannon entropy | 0.52356 |
| G+C content | 0.42567 |
| Mean single sequence MFE | -24.50 |
| Consensus MFE | -10.13 |
| Energy contribution | -12.63 |
| Covariance contribution | 2.50 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.636517 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 13190050 99 + 23011544 CUGUCUGGAUGUGGACAAGUUCCCACUUCUCCAACGACAGAUU-UCCAUUCGCACGCAAUGAAGAU-UAAGAACUUUAAGGCAUGCAAGGAACUAAAAAGU (((((((((.((((........))))...))))..)))))..(-(((.((.(((.((..(((((..-......)))))..)).)))))))))......... ( -27.10, z-score = -2.41, R) >droMoj3.scaffold_6500 12519242 82 + 32352404 -------------------UCUUUUGAAUAGAGACAACAUCUUGCUUAUCGAUAUGAAAUCAAAUUGUAACAAUUUCAAGGCUCACCAGGAACCCACCAGG -------------------...........(((........(((.(((.((((.((....)).))))))))))........))).((.((......)).)) ( -8.19, z-score = 1.18, R) >droSim1.chr2L 12984624 98 + 22036055 CUGUCUGGAUGUGGACGAGUUUCCACUUCUCCAACGACAGAUU-UCCAUUCGCACGCAAUGAAGAU-UAAGAACUUUAAGGCAUGCAAGAAACUAAAAAG- (((((((((.(((((......)))))...))))..)))))...-.......(((.((..(((((..-......)))))..)).)))..............- ( -26.10, z-score = -2.57, R) >droSec1.super_16 1344719 99 + 1878335 CUGUCUGGAUGUGGACGAGUUUCCACUUCUCCAACGACAGAUU-UCCAUUCGCACGCAAUGAAGAU-UAAGAACUUUAAGGCAUGCAAGGAACUAAAAAGU (((((((((.(((((......)))))...))))..)))))..(-(((.((.(((.((..(((((..-......)))))..)).)))))))))......... ( -28.40, z-score = -2.63, R) >droYak2.chr2L 9613625 99 + 22324452 CUGUCUGGAUGUGAAGGAGUUGCCUCUUCCCCAACGCCAGAUU-UCCAUUCGCACGCAAUGAAGAU-UAAGAACUUUAGGGCAUGCUGGGAACUACAAUAU ..((((((...((..((((......))))..))...))))))(-((((...(((.((..(((((..-......)))))..)).)))))))).......... ( -26.50, z-score = -0.96, R) >droEre2.scaffold_4929 14390817 99 - 26641161 CUGUCUGGAUGUGGAGGAGUUUCCUCUUCUCCAACGCCAGUUU-UUCAUUCGCACGCAAUGAAGAU-UAAGAACUUUAAGGCAUGCAACGUACUACUACGU ....((((...((((((((......))))))))...))))...-.......(((.((..(((((..-......)))))..)).))).(((((....))))) ( -30.70, z-score = -2.92, R) >consensus CUGUCUGGAUGUGGACGAGUUUCCACUUCUCCAACGACAGAUU_UCCAUUCGCACGCAAUGAAGAU_UAAGAACUUUAAGGCAUGCAAGGAACUAAAAAGU ..(((((....((((.(((......))).))))....))))).........(((.((..(((((.........)))))..)).)))............... (-10.13 = -12.63 + 2.50)

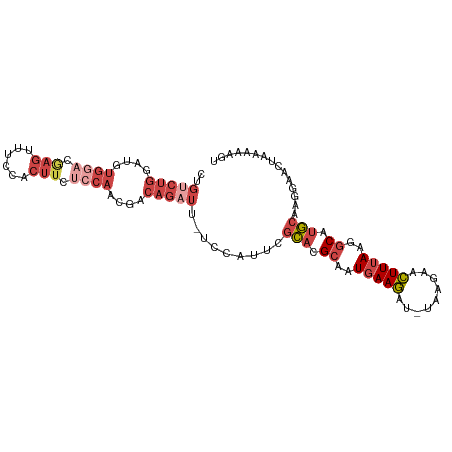
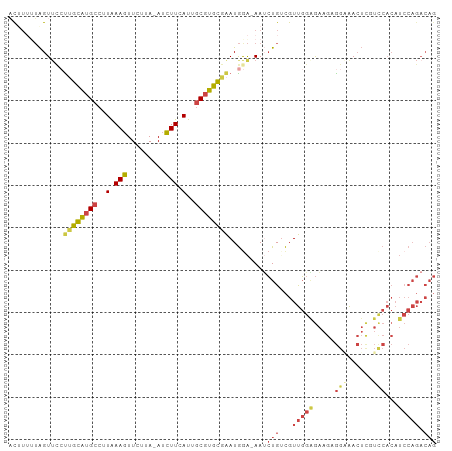
| Location | 13,190,050 – 13,190,149 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 72.91 |
| Shannon entropy | 0.52356 |
| G+C content | 0.42567 |
| Mean single sequence MFE | -26.57 |
| Consensus MFE | -9.90 |
| Energy contribution | -11.10 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.668239 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
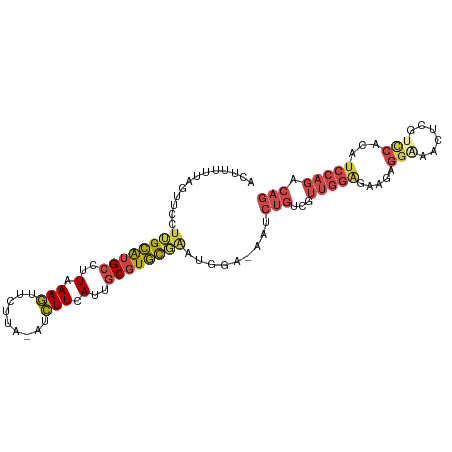
>dm3.chr2L 13190050 99 - 23011544 ACUUUUUAGUUCCUUGCAUGCCUUAAAGUUCUUA-AUCUUCAUUGCGUGCGAAUGGA-AAUCUGUCGUUGGAGAAGUGGGAACUUGUCCACAUCCAGACAG .........((((((((((((..(.(((......-..))).)..))))))))..)))-)..(((((..((((...(((((......))))).))))))))) ( -29.50, z-score = -2.60, R) >droMoj3.scaffold_6500 12519242 82 - 32352404 CCUGGUGGGUUCCUGGUGAGCCUUGAAAUUGUUACAAUUUGAUUUCAUAUCGAUAAGCAAGAUGUUGUCUCUAUUCAAAAGA------------------- ...(((((((((.....))))).((((((((........))))))))))))(((((........))))).............------------------- ( -12.80, z-score = 1.10, R) >droSim1.chr2L 12984624 98 - 22036055 -CUUUUUAGUUUCUUGCAUGCCUUAAAGUUCUUA-AUCUUCAUUGCGUGCGAAUGGA-AAUCUGUCGUUGGAGAAGUGGAAACUCGUCCACAUCCAGACAG -.......(((((((((((((..(.(((......-..))).)..)))))))...)))-)))(((((..((((...(((((......))))).))))))))) ( -28.70, z-score = -2.83, R) >droSec1.super_16 1344719 99 - 1878335 ACUUUUUAGUUCCUUGCAUGCCUUAAAGUUCUUA-AUCUUCAUUGCGUGCGAAUGGA-AAUCUGUCGUUGGAGAAGUGGAAACUCGUCCACAUCCAGACAG .........((((((((((((..(.(((......-..))).)..))))))))..)))-)..(((((..((((...(((((......))))).))))))))) ( -30.40, z-score = -3.28, R) >droYak2.chr2L 9613625 99 - 22324452 AUAUUGUAGUUCCCAGCAUGCCCUAAAGUUCUUA-AUCUUCAUUGCGUGCGAAUGGA-AAUCUGGCGUUGGGGAAGAGGCAACUCCUUCACAUCCAGACAG .........((((..((((((....(((......-..)))....))))))....)))-).(((((...((.(((.(((....))).))).)).)))))... ( -27.50, z-score = -1.28, R) >droEre2.scaffold_4929 14390817 99 + 26641161 ACGUAGUAGUACGUUGCAUGCCUUAAAGUUCUUA-AUCUUCAUUGCGUGCGAAUGAA-AAACUGGCGUUGGAGAAGAGGAAACUCCUCCACAUCCAGACAG .((((....))))((((((((..(.(((......-..))).)..)))))))).....-...((((...(((((..(((....))))))))...)))).... ( -30.50, z-score = -2.92, R) >consensus ACUUUUUAGUUCCUUGCAUGCCUUAAAGUUCUUA_AUCUUCAUUGCGUGCGAAUGGA_AAUCUGUCGUUGGAGAAGAGGAAACUCGUCCACAUCCAGACAG .............((((((((..(.(((.........))).)..)))))))).........(((...(((((.....(((......)))...))))).))) ( -9.90 = -11.10 + 1.20)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:37:11 2011