
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 13,173,536 – 13,173,633 |
| Length | 97 |
| Max. P | 0.582721 |

| Location | 13,173,536 – 13,173,633 |
|---|---|
| Length | 97 |
| Sequences | 13 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 71.31 |
| Shannon entropy | 0.57121 |
| G+C content | 0.58221 |
| Mean single sequence MFE | -37.57 |
| Consensus MFE | -14.16 |
| Energy contribution | -14.55 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.54 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.582721 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 13173536 97 - 23011544 GGCGAACCCAUGUCAAUG-GGCGGCGACGCAGCUGCCAAUAAU--GUACAAAUGG-CAGCAGAUGAAACAAUUGCUGCGUUAAACGGCGGAGCAGUGGGCG----------------- (.((..(((((....)))-)))).)..(((.(((((((.....--.......)))-))))........(.((((((.(((......))).)))))).))))----------------- ( -35.70, z-score = -1.70, R) >droSim1.chr2L 12968189 97 - 22036055 GGCGAACCCAUGUCGCUG-GGCGGCGACGCAGCUGCCAAUAAU--GUACAAAUGG-CAGCAGAUGAAACAAUUGCUGCGUUAAACGGCGGAGCAGUGGGCG----------------- ......(((((.((((((-((((((......))))))......--.........(-((((((.((...)).)))))))......))))))....)))))..----------------- ( -38.70, z-score = -1.94, R) >droSec1.super_16 1328321 97 - 1878335 GGCGAACCCAUGUCGCUG-GGCGGCGACGCAGCUGCCAAUAAU--GUACAAAUGG-CAGCAGAUGAAACAAUUGCUGCGUUAAACGGCGGAGCAGUGGGCG----------------- ......(((((.((((((-((((((......))))))......--.........(-((((((.((...)).)))))))......))))))....)))))..----------------- ( -38.70, z-score = -1.94, R) >droYak2.chr2L 9597085 97 - 22324452 GGCGAACCCAUGUCUCUA-GGCGGCGAUGCAGCUGCCAAUAAU--GUACAAAUGG-CAGCAGAUGAAACAAUUGCUGCGUUGAACGGCGGAGCUGUGGGAG----------------- ......((((((.((((.-((((((......)))))).....(--((.(((...(-((((((.((...)).))))))).))).)))..)))).))))))..----------------- ( -37.30, z-score = -2.39, R) >droEre2.scaffold_4929 14374272 97 + 26641161 GGCGAACCCAUGUCGCUA-GGCGGCGAUGCAGCUGCCACUAAU--GUACAAAUGG-CAGCAGAUGAAACAAUUGCUGCGUUGAACGGCGGAGCAGUGGGAG----------------- ......(((((.(((((.-((((((......))))))......--((.(((...(-((((((.((...)).))))))).))).)))))))....)))))..----------------- ( -38.70, z-score = -2.48, R) >droAna3.scaffold_12916 3908869 94 + 16180835 GGAGAGCCUAUGUCCCUC-GGCGGCGAGGCGGCCGCCAAUAAC--AUUCAAACGU-CGGCCGUGGAAACAAUUUCCGCUCUGAACGG---AUCGGUGGGCG----------------- .....((((((((((...-((((((......))))))......--.........(-(((..((((((.....)))))).))))..))---))..)))))).----------------- ( -36.30, z-score = -0.83, R) >droPer1.super_46 421164 97 + 590583 GGAGAGCCCAUGUCUUUG-GGCGGUGAUGCCGCCGCCCAUAAU--GUACAGAUGG-CCGCCGAUGAGACCAUUGCGGCCUUAAAUGGUGCUGCCGUGGGCG----------------- .....(((((((....((-(((((((....)))))))))....--((((....((-((((.((((....)))))))))).......))))...))))))).----------------- ( -49.40, z-score = -4.13, R) >droGri2.scaffold_15252 10204114 97 - 17193109 GGAGAGCCGAUGUCGCUG-GGCGGCGAUGCGGCCGCCAACAAU--GUGCAGAUGG-CAACGGACGAGACAAUUGCGGCGCUAAAUGGCGGAGCCGUUGGAG----------------- ..((.((((.((((.(((-((((((......))))))......--.(((.....)-)).)))....))))....)))).)).((((((...))))))....----------------- ( -36.00, z-score = -0.31, R) >droVir3.scaffold_12963 7050055 97 - 20206255 GGGGAGCCCAUGUCACUG-GGCGGCGAUGCUGCCGCCAACAAU--GUGCAGAUGU-CCACGGAUGAGACAAUUGCGGCACUGAAUGGCGGCGCAGUUGGGG----------------- .....(((((......))-)))..((((((.(((((((.((.(--((((((.(((-(((....)).)))).)))).))).))..)))))))))).)))...----------------- ( -39.00, z-score = -0.74, R) >droMoj3.scaffold_6500 26241160 97 - 32352404 GGGGAGCCCAUGUCGCUG-GGCGGGGAUGCAGCUGCCAAUAAU--GUGCAAAUGG-CCACGGAGGAGACAAUUGCAGCACUGAAUGGAGCUGCCGUUGGCG----------------- .((.(((((((.(((.((-(((((........)))))......--.(((((.((.-((.....))...)).))))).)).))))))).))).)).......----------------- ( -31.50, z-score = 1.10, R) >droWil1.scaffold_180708 6276399 97 - 12563649 GGUCAGCCAAUGUCUUUG-GGCGGCGAUGCCGCUGCCAAUACA--AUACAAAUGG-CCACCGAUGAGACGAUAGCCGCAUUGAAUGGUGGCGAUGUAGGCG----------------- .....(((.(((((....-(((((((....)))))))......--.........(-((((((..((..((.....))..))...)))))))))))).))).----------------- ( -35.60, z-score = -1.40, R) >dp4.chr4_group4 892049 97 - 6586962 GGAGAGCCCAUGUCUUUG-GGCGGUGAUGCCGCCGCCCAUAAU--GUACAGAUGG-CCGCCGAUGAGACCAUUGCGGCCUUAAAUGGUGCUGCCGUGGGCG----------------- .....(((((((....((-(((((((....)))))))))....--((((....((-((((.((((....)))))))))).......))))...))))))).----------------- ( -49.40, z-score = -4.13, R) >anoGam1.chr3R 51372520 116 - 53272125 ACCAAACGUGUGCCGUUACAGCAACUGCACCAGCAACAAGAACAAGUACAAACAAUCGGUUAUCGAUUCAA--GAUGUCUGGAACAUCUCUUACCCAACCGAAUACUAACAGUGGCGG ............(((((((......(((....)))....................((((((.........(--(((((.....)))))).......)))))).........))))))) ( -22.09, z-score = 0.13, R) >consensus GGCGAGCCCAUGUCGCUG_GGCGGCGAUGCAGCUGCCAAUAAU__GUACAAAUGG_CAGCAGAUGAAACAAUUGCUGCGUUGAACGGCGGAGCAGUGGGCG_________________ ......(((((........((((((......)))))).......................................((........))......)))))................... (-14.16 = -14.55 + 0.40)

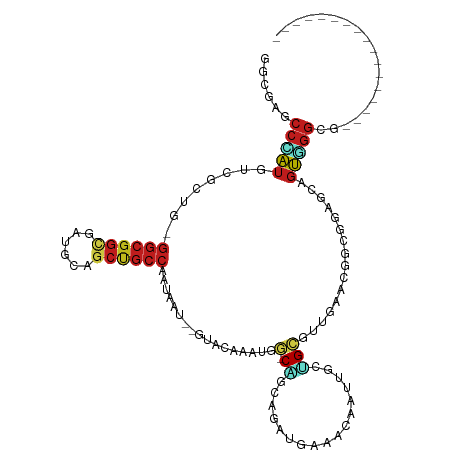
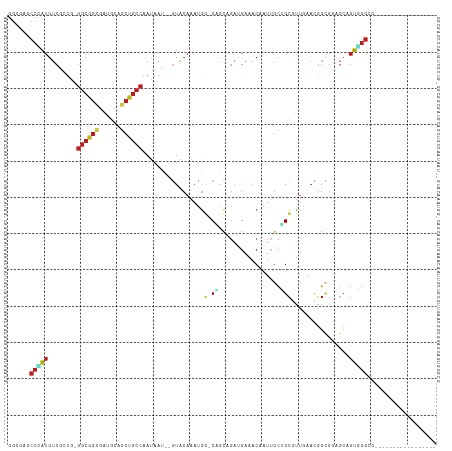
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:37:08 2011