| Sequence ID | dm3.chr2L |
|---|---|
| Location | 13,151,581 – 13,151,707 |
| Length | 126 |
| Max. P | 0.941091 |

| Location | 13,151,581 – 13,151,707 |
|---|---|
| Length | 126 |
| Sequences | 7 |
| Columns | 128 |
| Reading direction | forward |
| Mean pairwise identity | 68.77 |
| Shannon entropy | 0.62971 |
| G+C content | 0.51952 |
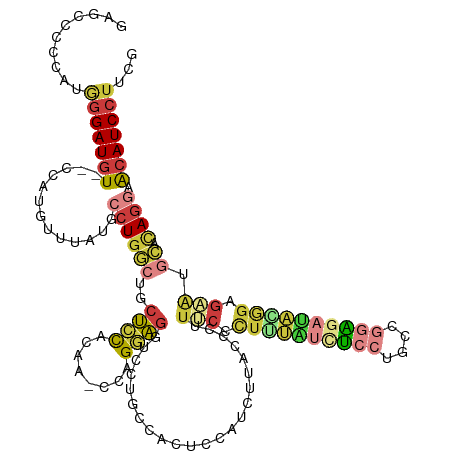
| Mean single sequence MFE | -35.60 |
| Consensus MFE | -19.97 |
| Energy contribution | -21.17 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.63 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.941091 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
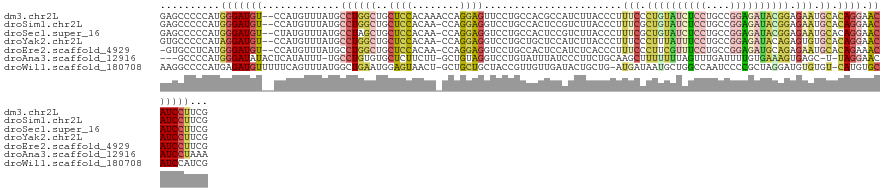
>dm3.chr2L 13151581 126 + 23011544 GAGCCCCCAUGGGAUGU--CCAUGUUUAUGCCUGGCUGCUCCACAAACCAGGAGUUCCUGCCACGCCAUCUUACCCUUUCCCUGUAUCUCCUGCCGGAGAUACGGAGAAUGCACAGGAACAUCCUUCG ..........(((((((--((.(((.(((((.((((.(((((........)))))....)))).))............((.((((((((((....)))))))))).))))).))))).)))))))... ( -46.00, z-score = -3.99, R) >droSim1.chr2L 12945798 125 + 22036055 GAGCCCCCAUGGGAUGU--CCAUGUUUAUGCCUGGCUGCUCCACAA-CCAGGAGGUCCUGCCACUCCGUCUUACCCUUUCGCUGUAUCUCCUGCCGGAGAUACGGAGAAUGCACAGGAACAUCCUUCG ..........(((((((--.(((....)))((((((..((((....-...((((.........))))................((((((((....))))))))))))...)).)))).)))))))... ( -41.20, z-score = -1.53, R) >droSec1.super_16 1306606 125 + 1878335 GAGCCCCCAUGGGAUGU--CUAUGUUUAUGCCUAGCUGCUCCACAA-CCAGGAGGUCCUGCCACUCCGUCUUACCCUUUCGCUGUAUCUCCUGCCGGAGAUACGGAGAAUGCACAGGAACAUCCUUCG ..........(((((((--...........(((.((..((((....-...((((.........))))................((((((((....))))))))))))...))..))).)))))))... ( -36.20, z-score = -0.74, R) >droYak2.chr2L 9574886 125 + 22324452 GUGCCCCCAUAGGAUGU--CCAUGUUUAUGCCUGGCUGCUCCACAA-CCAGGAGGUCCUGCUGCUCCAUCUUACCCUUUCCCUUUAUUUCCUGCCGGAGAUACAGAGUGUGCACAGGAACAUCCUUCG ..........(((((((--((.(((.(((((((((..((..((..(-((....)))..))..))..).................(((((((....)))))))))).))))).))))).)))))))... ( -33.30, z-score = -0.61, R) >droEre2.scaffold_4929 14352360 124 - 26641161 -GUGCCUCAUGGGAUGU--CCAUGUUUAUGCCUGGCUGCUCCACAA-CCAGGAGGUCCUGCCACUCCAUCUCACCCUUUCCCUUCGUUUCCUGCCGGAGAUGCAGAGAAUGCACAGAAACAUCCUUCG -((((....(((((((.--...........(((((.((.....)).-))))).((.....))....)))))))....(((.((.(((((((....))))))).)).))).)))).(((......))). ( -34.70, z-score = -0.65, R) >droAna3.scaffold_12916 3890480 121 - 16180835 ---GCCCCAUGGGAUAUACUCAUAUUU-UGCCUGUGUGCUCUUCUU-GCUGUAGGUCCUGUAUUUAUCCCUUCUGCAAGCUUUUUUUAGUUUGAUUUUGUGAAAGUGAGC-U-UAGGAACAUCCUAAA ---((..(((((((((.....((((..-.(((((...((.......-))..)))))...)))).)))))).....((((((......))))))...........))).))-(-((((.....))))). ( -26.80, z-score = -0.87, R) >droWil1.scaffold_180708 6247224 125 + 12563649 AAGGCCCCAUGAGAUGUUUUUCAGUUUAUGGCUGAAUGGAGUAACU-GCUGCUGCUACCGUUGUUGAUACUGCUG-AUGAUAAUGCUGGCCAAUCCCCGCUAGGAUGUGUGU-CAUGUGCAUCCAUCG ..(((((((((((.((.....)).)))))))...(((((((((.(.-...).)))).))))).............-...........))))...........(((((..(..-...)..))))).... ( -31.00, z-score = 0.56, R) >consensus GAGCCCCCAUGGGAUGU__CCAUGUUUAUGCCUGGCUGCUCCACAA_CCAGGAGGUCCUGCCACUCCAUCUUACCCUUUCCCUUUAUCUCCUGCCGGAGAUACGGAGAAUGCACAGGAACAUCCUUCG ..........(((((((.............((((((..((((........)))).......................(((.((((((((((....)))))))))).))).)).)))).)))))))... (-19.97 = -21.17 + 1.20)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:37:06 2011