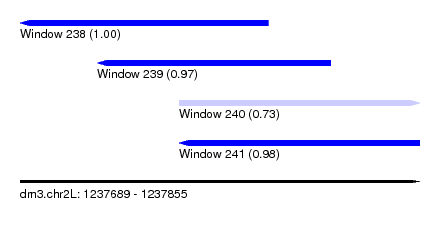
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 1,237,689 – 1,237,855 |
| Length | 166 |
| Max. P | 0.997435 |

| Location | 1,237,689 – 1,237,792 |
|---|---|
| Length | 103 |
| Sequences | 8 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 90.94 |
| Shannon entropy | 0.17786 |
| G+C content | 0.39091 |
| Mean single sequence MFE | -30.82 |
| Consensus MFE | -27.55 |
| Energy contribution | -27.60 |
| Covariance contribution | 0.05 |
| Combinations/Pair | 1.13 |
| Mean z-score | -3.38 |
| Structure conservation index | 0.89 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.10 |
| SVM RNA-class probability | 0.997435 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 1237689 103 - 23011544 GCGUUCGUGGAAAUUAGCGGCAGCUUCCCAGCUGCUGCUGAUGCGAUUUUGAUGUGCAAAAUAUUAAUUAUAAAACAAUAAUUGAAUUGUGCUCUUGCUCUUG------ (((.((((....(((((((((((((....)))))))))))))))))....((.(..(((....((((((((......)))))))).)))..))).))).....------ ( -32.50, z-score = -3.46, R) >droSim1.chr2L 1209861 103 - 22036055 GCGUUCGUGGAAAUUAGCGGCAGCUUCCCAGCUGCUGCUGAUGCGAUUUUGAUGUGCAAAAUAUUAAUUAUAAAACAAUAAUUGAAUUGUGCUCUUGCUCUUG------ (((.((((....(((((((((((((....)))))))))))))))))....((.(..(((....((((((((......)))))))).)))..))).))).....------ ( -32.50, z-score = -3.46, R) >droSec1.super_14 1188632 103 - 2068291 GCGUUCGUGGAAAUUAGCGGCAGCUUCCCAGCUGCUGCUGAUGCGAUUUUGAUGUGCAAAAUAUUAAUUAUAAAACAAUAAUUGAAUUGUGCUCUUGCUCUUG------ (((.((((....(((((((((((((....)))))))))))))))))....((.(..(((....((((((((......)))))))).)))..))).))).....------ ( -32.50, z-score = -3.46, R) >droYak2.chr2L 1212631 109 - 22324452 GCGUUGGUGGAAAUUAGCGGCAGCUUCCCAGCUGCUGCUGAUGCGAUUUUGAUGUGCAAAAUAUUAAUUAUAAAACAAUAAUUGAAUUGUGCUCUUGCUCUUGCUCUUG (((..((..(..(((((((((((((....)))))))))))))........((.(..(((....((((((((......)))))))).)))..))))..))..)))..... ( -34.70, z-score = -3.40, R) >droEre2.scaffold_4929 1279306 103 - 26641161 GCGCUCGUGGAAAUUAGCAGCAGCUUCCCAGCUGCUGCUGAUGCGAUUUUGAUGUGCAAAAUAUUAAUUAUAAAACAAUAAUUGAAUUGUGCUCUUGCUCUUG------ ((..((((....(((((((((((((....)))))))))))))))))....((.(..(((....((((((((......)))))))).)))..)))..)).....------ ( -33.10, z-score = -3.54, R) >droAna3.scaffold_12916 4699276 103 - 16180835 GCCUCCGUGGAAAUUAGUGGCAGCUUCCCAGCUGCUGCUGAUGCGAUUUUGAUGUGCAAAAUAUUAAUUAUAAAACAAUAAUUGAAUUGUGCUCUUGUUCGUG------ .((.....))..(((((..((((((....))))))..)))))((((....((.(..(((....((((((((......)))))))).)))..)))....)))).------ ( -31.70, z-score = -3.90, R) >dp4.chr4_group3 9696048 90 - 11692001 -------UGGAAAUUAGAGGCAGCUUCCCAGCUGCCGCUGAUGCGAUUUUGAUGUGCAAAAUAUUAAUUAUAAAACAAUAAUUGAAUUGUGCUCUUG------------ -------.....(((((.(((((((....))))))).)))))........((.(..(((....((((((((......)))))))).)))..)))...------------ ( -24.80, z-score = -2.90, R) >droPer1.super_8 894578 90 - 3966273 -------UGGAAAUUAGAGGCAGCUUCCCAGCUGCCGCUGAUGCGAUUUUGAUGUGCAAAAUAUUAAUUAUAAAACAAUAAUUGAAUUGUGCUCUUG------------ -------.....(((((.(((((((....))))))).)))))........((.(..(((....((((((((......)))))))).)))..)))...------------ ( -24.80, z-score = -2.90, R) >consensus GCGUUCGUGGAAAUUAGCGGCAGCUUCCCAGCUGCUGCUGAUGCGAUUUUGAUGUGCAAAAUAUUAAUUAUAAAACAAUAAUUGAAUUGUGCUCUUGCUCUUG______ ............(((((((((((((....)))))))))))))((((....((.(..(((....((((((((......)))))))).)))..)))))))........... (-27.55 = -27.60 + 0.05)
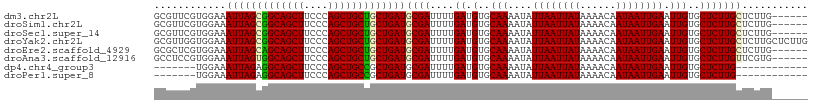
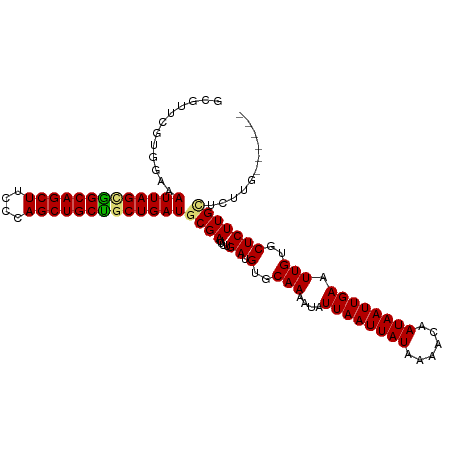
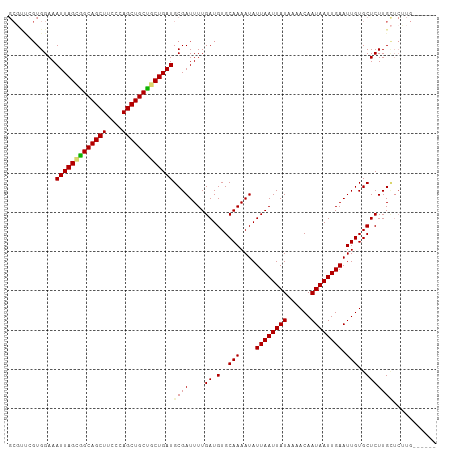
| Location | 1,237,721 – 1,237,818 |
|---|---|
| Length | 97 |
| Sequences | 8 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 88.35 |
| Shannon entropy | 0.23900 |
| G+C content | 0.48692 |
| Mean single sequence MFE | -34.49 |
| Consensus MFE | -25.97 |
| Energy contribution | -25.79 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.88 |
| SVM RNA-class probability | 0.973059 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 1237721 97 - 23011544 GUGCCACGCCCAUAAGUGCGUGGGAGGCGUUCGUGGAAAUUAGCGGCAGCUUCCCAGCUGCUGCUGAUGCGAUUUUGAUGUGCAAAAUAUUAAUUAU ..(((...(((((......))))).)))..((((....(((((((((((((....)))))))))))))))))..(((((((.....))))))).... ( -37.60, z-score = -2.69, R) >droSim1.chr2L 1209893 97 - 22036055 GUGCCACGCCCAUAAGUGCGUGGGAGGCGUUCGUGGAAAUUAGCGGCAGCUUCCCAGCUGCUGCUGAUGCGAUUUUGAUGUGCAAAAUAUUAAUUAU ..(((...(((((......))))).)))..((((....(((((((((((((....)))))))))))))))))..(((((((.....))))))).... ( -37.60, z-score = -2.69, R) >droSec1.super_14 1188664 97 - 2068291 GUGCCACGCCCAUAAGUGCGUGGGAGGCGUUCGUGGAAAUUAGCGGCAGCUUCCCAGCUGCUGCUGAUGCGAUUUUGAUGUGCAAAAUAUUAAUUAU ..(((...(((((......))))).)))..((((....(((((((((((((....)))))))))))))))))..(((((((.....))))))).... ( -37.60, z-score = -2.69, R) >droYak2.chr2L 1212669 97 - 22324452 GUGCCACGCCCAUAAGUGCGUGGGAGGCGUUGGUGGAAAUUAGCGGCAGCUUCCCAGCUGCUGCUGAUGCGAUUUUGAUGUGCAAAAUAUUAAUUAU ...((((((.(....).))))))...((((..(.....(((((((((((((....)))))))))))))......)..))))................ ( -36.40, z-score = -2.14, R) >droEre2.scaffold_4929 1279338 97 - 26641161 GUGCCACGCCCAUAAGUGCGUGGUGGGCGCUCGUGGAAAUUAGCAGCAGCUUCCCAGCUGCUGCUGAUGCGAUUUUGAUGUGCAAAAUAUUAAUUAU (((((.(((((........).)))))))))((((....(((((((((((((....)))))))))))))))))..(((((((.....))))))).... ( -40.30, z-score = -3.40, R) >droAna3.scaffold_12916 4699308 96 - 16180835 GUGCCACACCCAUAAGU-CGACGAACGCCUCCGUGGAAAUUAGUGGCAGCUUCCCAGCUGCUGCUGAUGCGAUUUUGAUGUGCAAAAUAUUAAUUAU .(((.(((..((.((((-(((((........)))....(((((..((((((....))))))..))))).)))))))).))))))............. ( -28.80, z-score = -2.56, R) >dp4.chr4_group3 9696074 84 - 11692001 GCGCUGUGCCCAUGAGUGC-------------GUGGAAAUUAGAGGCAGCUUCCCAGCUGCCGCUGAUGCGAUUUUGAUGUGCAAAAUAUUAAUUAU .(((.....(((((....)-------------))))..(((((.(((((((....))))))).))))))))...(((((((.....))))))).... ( -28.80, z-score = -2.45, R) >droPer1.super_8 894604 84 - 3966273 GCGCUGUGCCCAUGAGUGC-------------GUGGAAAUUAGAGGCAGCUUCCCAGCUGCCGCUGAUGCGAUUUUGAUGUGCAAAAUAUUAAUUAU .(((.....(((((....)-------------))))..(((((.(((((((....))))))).))))))))...(((((((.....))))))).... ( -28.80, z-score = -2.45, R) >consensus GUGCCACGCCCAUAAGUGCGUGGGAGGCGUUCGUGGAAAUUAGCGGCAGCUUCCCAGCUGCUGCUGAUGCGAUUUUGAUGUGCAAAAUAUUAAUUAU ......(((((((...................))))..(((((((((((((....))))))))))))))))...(((((((.....))))))).... (-25.97 = -25.79 + -0.19)

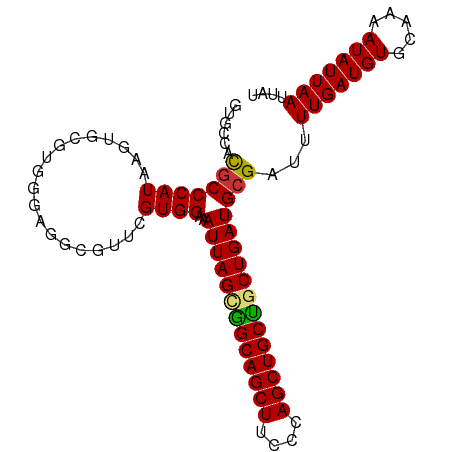
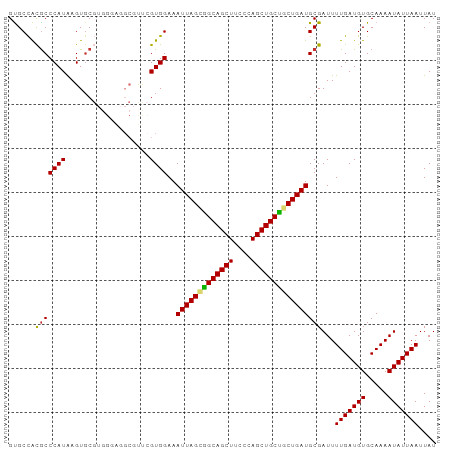
| Location | 1,237,755 – 1,237,855 |
|---|---|
| Length | 100 |
| Sequences | 7 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 73.83 |
| Shannon entropy | 0.54184 |
| G+C content | 0.59636 |
| Mean single sequence MFE | -34.87 |
| Consensus MFE | -19.27 |
| Energy contribution | -19.84 |
| Covariance contribution | 0.57 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.10 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.732669 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 1237755 100 + 23011544 CAGCAGCUGGGAAGCUGCCGCUAAUUUCCACGAACGCCUCCCACGCACUUAUGGGCGUGGCACACGCACAUUCACUGGUUGAUGGAGGGGGGGAGUUGGG ..((((((....))))))..(((((((((.....(.((((((((((.(....).)))))((....))................))))).)))))))))). ( -37.20, z-score = -0.97, R) >droSim1.chr2L 1209927 100 + 22036055 CAGCAGCUGGGAAGCUGCCGCUAAUUUCCACGAACGCCUCCCACGCACUUAUGGGCGUGGCACACGCACAUUCGCUGGUUGAUGGAAAGGGGGAGCUGAG ((((((((....)))).((.((..((((((..(((.....((((((.(....).))))))..((.((......)))))))..)))))))).)).)))).. ( -34.20, z-score = -0.25, R) >droSec1.super_14 1188698 100 + 2068291 CAGCAGCUGGGAAGCUGCCGCUAAUUUCCACGAACGCCUCCCACGCACUUAUGGGCGUGGCACACGCACAUUCGCCGGUUGAUGGAAGGGGGGAGUUGGG ..((((((....))))))............(.(((.((((((...((...((.((((((((....)).))..)))).))...))...)))))).))).). ( -35.90, z-score = -0.18, R) >droYak2.chr2L 1212703 99 + 22324452 CAGCAGCUGGGAAGCUGCCGCUAAUUUCCACCAACGCCUCCCACGCACUUAUGGGCGUGGCACACGCA-CAUUCAAAGGUGGGGGAGGGGGGCAGUUGGA ..((((((....))))))............((((((((((((.(.((((((((.(((((...))))).-)))....))))).)...))))))).))))). ( -45.20, z-score = -2.89, R) >droEre2.scaffold_4929 1279372 87 + 26641161 CAGCAGCUGGGAAGCUGCUGCUAAUUUCCACGAGCGCCCACCACGCACUUAUGGGCGUGGCACACGCACAUGCAAAGGUUCUUGGGA------------- ((((((((....))))))))......(((.((((.(((..((((((.(....).)))))).....((....))...))).)))))))------------- ( -36.20, z-score = -2.35, R) >droAna3.scaffold_12916 4699342 97 + 16180835 CAGCAGCUGGGAAGCUGCCACUAAUUUCCACGGAGGCGUUCGUCG-ACUUAUGGGUGUGGCACACGCA-AUUC-UUAGUAUAUGGCAGGGAAAAUGAGGA ..((((((....))))))........(((......((((..((((-(((....))).))))..)))).-.(((-(..((.....))..)))).....))) ( -25.40, z-score = 0.42, R) >droPer1.super_8 894638 75 + 3966273 CGGCAGCUGGGAAGCUGCCUCUAAUUUCCAC-------------GCACUCAUGGGCACAGCGCACGCA-AGGCCUUGGGAGGAGGAGGA----------- .(((((((....))))))).....(((((.(-------------.(.((((.((((...((....)).-..)))))))).)).))))).----------- ( -30.00, z-score = -1.50, R) >consensus CAGCAGCUGGGAAGCUGCCGCUAAUUUCCACGAACGCCUCCCACGCACUUAUGGGCGUGGCACACGCACAUUCACUGGUUGAUGGAAGGGGGGAGUUGGG ..((((((....))))))......(((((............(((((.(....).)))))((....))................)))))............ (-19.27 = -19.84 + 0.57)
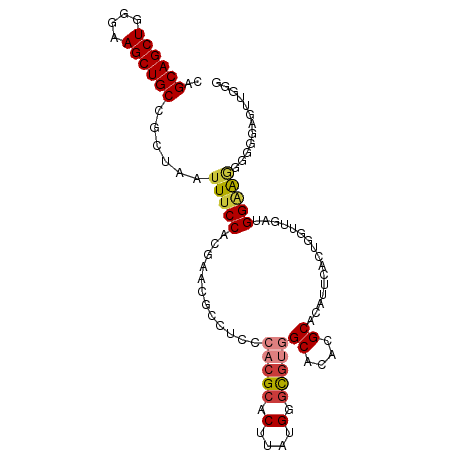
| Location | 1,237,755 – 1,237,855 |
|---|---|
| Length | 100 |
| Sequences | 7 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 73.83 |
| Shannon entropy | 0.54184 |
| G+C content | 0.59636 |
| Mean single sequence MFE | -32.69 |
| Consensus MFE | -20.59 |
| Energy contribution | -22.73 |
| Covariance contribution | 2.15 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.09 |
| SVM RNA-class probability | 0.981923 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 1237755 100 - 23011544 CCCAACUCCCCCCCUCCAUCAACCAGUGAAUGUGCGUGUGCCACGCCCAUAAGUGCGUGGGAGGCGUUCGUGGAAAUUAGCGGCAGCUUCCCAGCUGCUG ..............(((.(((.....)))(((.((((.(.((((((.(....).)))))).).)))).))))))......((((((((....)))))))) ( -34.10, z-score = -1.60, R) >droSim1.chr2L 1209927 100 - 22036055 CUCAGCUCCCCCUUUCCAUCAACCAGCGAAUGUGCGUGUGCCACGCCCAUAAGUGCGUGGGAGGCGUUCGUGGAAAUUAGCGGCAGCUUCCCAGCUGCUG ....(((.....(((((((.(((..(((....)))((.(.((((((.(....).)))))).).))))).)))))))..)))(((((((....))))))). ( -37.50, z-score = -1.92, R) >droSec1.super_14 1188698 100 - 2068291 CCCAACUCCCCCCUUCCAUCAACCGGCGAAUGUGCGUGUGCCACGCCCAUAAGUGCGUGGGAGGCGUUCGUGGAAAUUAGCGGCAGCUUCCCAGCUGCUG .............((((((.(((..(((....)))((.(.((((((.(....).)))))).).))))).)))))).....((((((((....)))))))) ( -35.60, z-score = -1.23, R) >droYak2.chr2L 1212703 99 - 22324452 UCCAACUGCCCCCCUCCCCCACCUUUGAAUG-UGCGUGUGCCACGCCCAUAAGUGCGUGGGAGGCGUUGGUGGAAAUUAGCGGCAGCUUCCCAGCUGCUG .....(..((.(((((((((((.((...(((-.(((((...))))).)))))))).).)))))).)..))..).......((((((((....)))))))) ( -37.70, z-score = -2.10, R) >droEre2.scaffold_4929 1279372 87 - 26641161 -------------UCCCAAGAACCUUUGCAUGUGCGUGUGCCACGCCCAUAAGUGCGUGGUGGGCGCUCGUGGAAAUUAGCAGCAGCUUCCCAGCUGCUG -------------...........(((.((((.((((.((((((((.(....).)))))))).)))).)))).)))....((((((((....)))))))) ( -39.90, z-score = -3.52, R) >droAna3.scaffold_12916 4699342 97 - 16180835 UCCUCAUUUUCCCUGCCAUAUACUAA-GAAU-UGCGUGUGCCACACCCAUAAGU-CGACGAACGCCUCCGUGGAAAUUAGUGGCAGCUUCCCAGCUGCUG .....................(((((-....-((.(((.....))).))....(-(.(((........))).))..)))))(((((((....))))))). ( -19.90, z-score = -0.31, R) >droPer1.super_8 894638 75 - 3966273 -----------UCCUCCUCCUCCCAAGGCCU-UGCGUGCGCUGUGCCCAUGAGUGC-------------GUGGAAAUUAGAGGCAGCUUCCCAGCUGCCG -----------(((.(...(((....(((..-.((....))...)))...)))...-------------).))).......(((((((....))))))). ( -24.10, z-score = -0.86, R) >consensus CCCAACUCCCCCCUUCCAUCAACCAGCGAAUGUGCGUGUGCCACGCCCAUAAGUGCGUGGGAGGCGUUCGUGGAAAUUAGCGGCAGCUUCCCAGCUGCUG ..............(((((..............((((.(.((((((.(....).)))))).).))))..)))))......((((((((....)))))))) (-20.59 = -22.73 + 2.15)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:08:00 2011