
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 13,148,451 – 13,148,551 |
| Length | 100 |
| Max. P | 0.579829 |

| Location | 13,148,451 – 13,148,551 |
|---|---|
| Length | 100 |
| Sequences | 9 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 74.85 |
| Shannon entropy | 0.47865 |
| G+C content | 0.40235 |
| Mean single sequence MFE | -25.49 |
| Consensus MFE | -14.74 |
| Energy contribution | -15.04 |
| Covariance contribution | 0.30 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.13 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.579829 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 13148451 100 - 23011544 CAUUUGAAGCUUGGCCUGGAAAG-CAAAAUAUUGUU----ACUGCGCCAAUAUGUUUUGC------CAGAUAUCU---AUCUUUAGUGGC----CCAGGCCAGAAUGUAUUAUGAUUU ..........(((((((((((((-((...(((((..----.......)))))))))))((------((..((...---.....)).))))----)))))))))............... ( -24.90, z-score = -0.85, R) >droWil1.scaffold_180708 6242972 108 - 12563649 --CAGUCAAGUUGGCCUGAAAACCCAAAAUAUUGUUACAACAGCCAGCAAUAUGUUGUGUUUGCUGCAGAUAUCUUAGAUAUCGUUAGUU----UUGGGCCAGA----GCUAUGAUUU --.(((((((((((((..(((((.(((.((((((((.........)))))))).))).))))((((..((((((...))))))..)))).----)..)))))..----))).))))). ( -30.90, z-score = -2.51, R) >droPer1.super_46 389339 100 + 590583 -----GAAGUUUGGCCUGAAAAACCAAAAUAUUGUU----ACUGCCGCAAUAUGUUUUGU------CAGAUAUUC---AUCUUUAGUGGCCAGGCCGGGCCAGAAUAUGUUACGAUUG -----...((((((((((.(((((....(((((((.----......))))))))))))((------((..((...---.....)).)))))))))))))).................. ( -22.80, z-score = -0.38, R) >dp4.chr4_group4 860551 100 - 6586962 -----GAAGUUUGGCCUGAAAAACCAAAAUAUUGUU----ACUGCCGCAAUAUGUUUUGU------CAGAUAUUC---AUCUUUAGUGGCCAGGCCGGGCCAGAAUAUGUUACGAUUG -----...((((((((((.(((((....(((((((.----......))))))))))))((------((..((...---.....)).)))))))))))))).................. ( -22.80, z-score = -0.38, R) >droAna3.scaffold_12916 3887883 88 + 16180835 CAAGGGAAACAAGGCCUGGAAA-ACAAAAUAUUGUU----CCUCCAACAAUAUGUUAUGU------CAGAUAUUU---AUCUCUAGAGUU----UCAGGUCCAU----AU-------- ....(....)..(((((((((.-.(...((((((((----.....)))))))).......------.((((....---))))...)..))----)))))))...----..-------- ( -19.10, z-score = -1.50, R) >droEre2.scaffold_4929 14349276 100 + 26641161 CAUUCGCAGCUUGGCCUGGAAAG-CAAAAUAUUGUU----ACUGCGCCAAUAUGUUUUGC------CUGAUAUCC---AUCUUUAGUGGC----CCAGGCCAGAAUGUGUUAUGAUUU (((.((((..(((((((((...(-((((((((.(((----........))))))))))))------.......((---((.....)))).----)))))))))..))))..))).... ( -32.70, z-score = -2.79, R) >droYak2.chr2L 9571626 100 - 22324452 CAUUUGGAGCCUGGCCUGGAAAG-CAAAAUAUUGUU----ACUGCGCCAAUAUGUUUUGC------CAGAUAUCC---AUCUUUAGUGGC----CCAGGCCAGAAUUUGUUAUGAUUU (((.(..(..(((((((((...(-((((((((.(((----........))))))))))))------.......((---((.....)))).----)))))))))..)..)..))).... ( -29.90, z-score = -1.85, R) >droSec1.super_16 1303487 100 - 1878335 CAUUUGAAGCUUGGCCUGGAAAG-CAAAAUAUUGUU----ACUGCGCCAAUAUGUUUUGC------CAGAUAUCC---AUCUUUAGUGGC----CCAGGCCAGAAUGUGUUAUGAUUU ..........(((((((((...(-((((((((.(((----........))))))))))))------.......((---((.....)))).----)))))))))............... ( -26.20, z-score = -1.02, R) >droSim1.chr2L 12942329 100 - 22036055 CAUUUGAAGCUUGGCCUGGAAAG-CAAAAUAUUGUU----ACUGCGCCAAUAUGUUUUGC------CAGAUAUCC---AUCUUUAGUGGC----CCAUGCCAGAAUGUGUUAUGGUUU (((((...((.(((.((((((((-((...(((((..----.......))))))))))).)------)))....((---((.....)))).----))).))..)))))........... ( -20.10, z-score = 1.08, R) >consensus CAUUUGAAGCUUGGCCUGGAAAG_CAAAAUAUUGUU____ACUGCGCCAAUAUGUUUUGC______CAGAUAUCC___AUCUUUAGUGGC____CCAGGCCAGAAUGUGUUAUGAUUU ..........(((((((((.....((((((((...................))))))))........((((.......))))............)))))))))............... (-14.74 = -15.04 + 0.30)

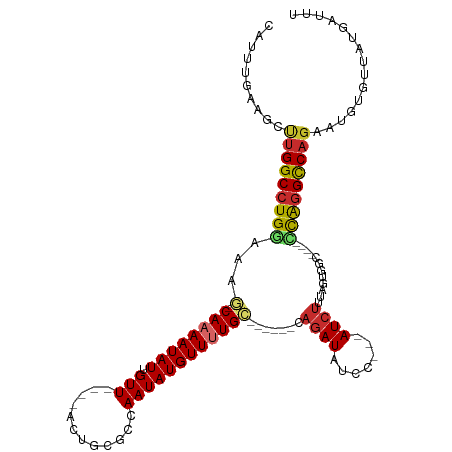

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:37:04 2011